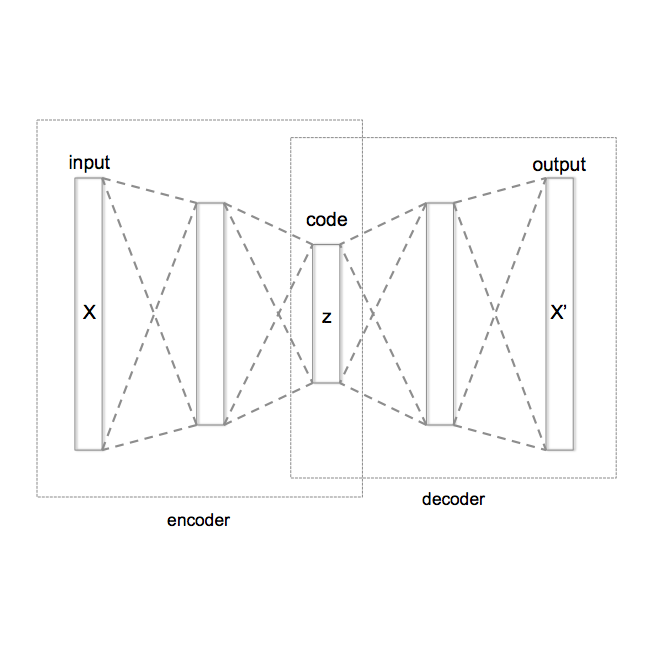
Purpose. Brain Magnetic Resonance Images (MRIs) are essential for the diagnosis of neurological diseases. Recently, deep learning methods for unsupervised anomaly detection (UAD) have been proposed for the analysis of brain MRI. These methods rely on healthy brain MRIs and eliminate the requirement of pixel-wise annotated data compared to supervised deep learning. While a wide range of methods for UAD have been proposed, these methods are mostly 2D and only learn from MRI slices, disregarding that brain lesions are inherently 3D and the spatial context of MRI volumes remains unexploited. Methods. We investigate whether using increased spatial context by using MRI volumes combined with spatial erasing leads to improved unsupervised anomaly segmentation performance compared to learning from slices. We evaluate and compare 2D variational autoencoder (VAE) to their 3D counterpart, propose 3D input erasing, and systemically study the impact of the data set size on the performance. Results. Using two publicly available segmentation data sets for evaluation, 3D VAE outperform their 2D counterpart, highlighting the advantage of volumetric context. Also, our 3D erasing methods allow for further performance improvements. Our best performing 3D VAE with input erasing leads to an average DICE score of 31.40% compared to 25.76% for the 2D VAE. Conclusions. We propose 3D deep learning methods for UAD in brain MRI combined with 3D erasing and demonstrate that 3D methods clearly outperform their 2D counterpart for anomaly segmentation. Also, our spatial erasing method allows for further performance improvements and reduces the requirement for large data sets.
翻译:脑磁共振成像(MRIs)是诊断神经疾病所必需的。最近,为分析大脑磁共振成像(UAD),提出了用于分析大脑磁共振成像(UAD)的深层学习方法。这些方法依靠健康的大脑光成像(MRIs),并消除了对像素的附加数据的要求,而没有监督深度学习。虽然提出了UAD的多种方法,但这些方法大多为2D,只从MRI切片中学习,无视大脑损伤本质上是3D,而磁共振量的空间环境环境仍然没有得到开发。方法。我们研究的是,使用磁共振成像(UAD)体积结合空间变异(UA)体积和空间变异(UA)法是否提高了不受监督的异常分解性性功能,我们评估和比较2D变异形自动变异变异数据(VE)到3D的3D值分析方法,我们用3D分解为3D的更精确的进化法,我们用3D的进化方法可以降低3A的进化方法。