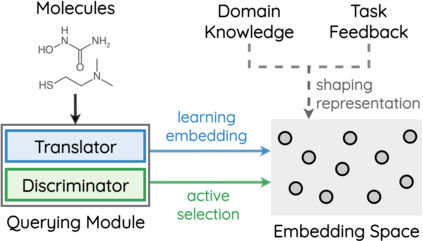
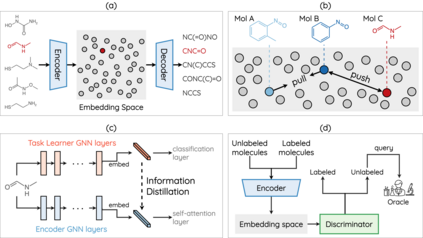
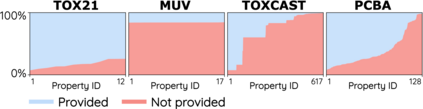
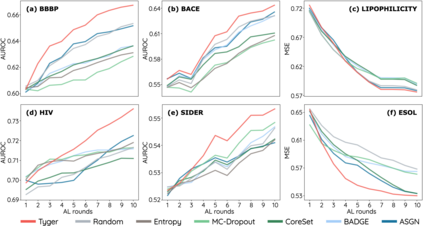
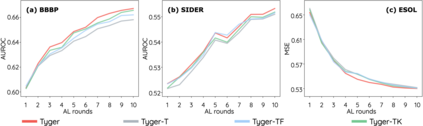
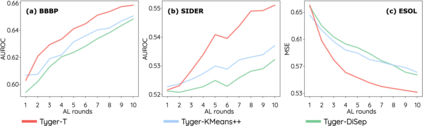
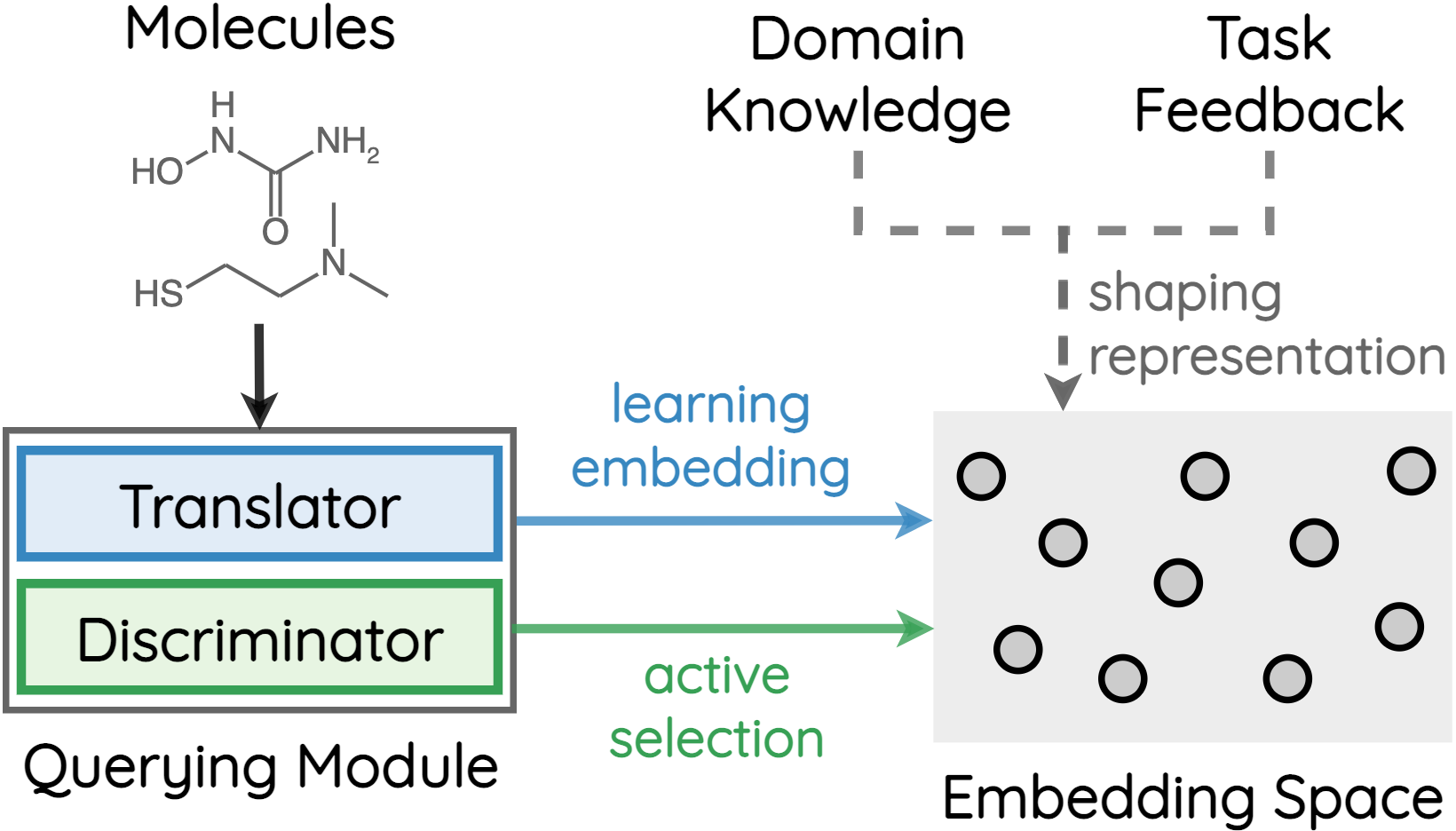
How to accurately predict the properties of molecules is an essential problem in AI-driven drug discovery, which generally requires a large amount of annotation for training deep learning models. Annotating molecules, however, is quite costly because it requires lab experiments conducted by experts. To reduce annotation cost, deep Active Learning (AL) methods are developed to select only the most representative and informative data for annotating. However, existing best deep AL methods are mostly developed for a single type of learning task (e.g., single-label classification), and hence may not perform well in molecular property prediction that involves various task types. In this paper, we propose a Task-type-generic active learning framework (termed Tyger) that is able to handle different types of learning tasks in a unified manner. The key is to learn a chemically-meaningful embedding space and perform active selection fully based on the embeddings, instead of relying on task-type-specific heuristics (e.g., class-wise prediction probability) as done in existing works. Specifically, for learning the embedding space, we instantiate a querying module that learns to translate molecule graphs into corresponding SMILES strings. Furthermore, to ensure that samples selected from the space are both representative and informative, we propose to shape the embedding space by two learning objectives, one based on domain knowledge and the other leveraging feedback from the task learner (i.e., model that performs the learning task at hand). We conduct extensive experiments on benchmark datasets of different task types. Experimental results show that Tyger consistently achieves high AL performance on molecular property prediction, outperforming baselines by a large margin. We also perform ablative experiments to verify the effectiveness of each component in Tyger.
翻译:如何准确预测分子特性是AI驱动的药物发现中的一个基本问题。 AI驱动的药物发现通常需要大量的批注来培训深层学习模型。 但是, 批注分子费用非常昂贵, 因为它需要专家进行实验室实验。 为了降低批注成本, 深度主动学习(AL) 方法只选择最有代表性、 信息最丰富的数据进行批注。 但是, 现有的最深的AL 方法大多是为单一类型的学习任务( 如单标签分类) 开发的, 因此可能无法在涉及不同任务类型的分子属性预测中产生良好的效果。 具体地说, 我们提议一个任务类型为型的分子实验性研究框架( 名为Tyger ), 因为它可以以统一的方式处理不同类型的学习任务。 关键在于学习一个具有化学意义的嵌入空间的嵌入空间空间模型, 并且完全根据嵌入式模型进行积极的选择, 而不是依赖特定任务型号模型( 例如, 等级预测概率概率) 。 具体地说, 学习嵌入空间的模型, 我们从一个任务, 运行一个, 运行一个, 运行一个任务 运行一个 运行一个任务到一个连续的模型, 运行一个任务 运行一个模块, 学习一个任务 学习一个任务, 学习一个任务 学习一个任务 运行中 学习一个任务 运行中 运行中 学习一个任务, 运行中 学习一个任务 学习一个任务到一个任务到一个任务 运行成一个任务 学习一个任务 运行成一个任务 。