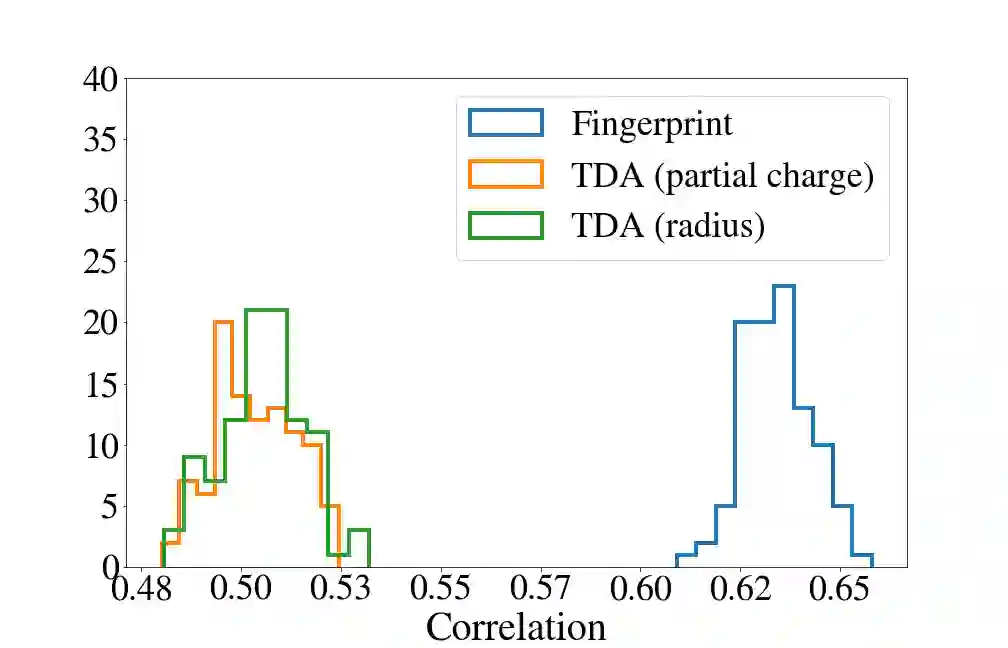
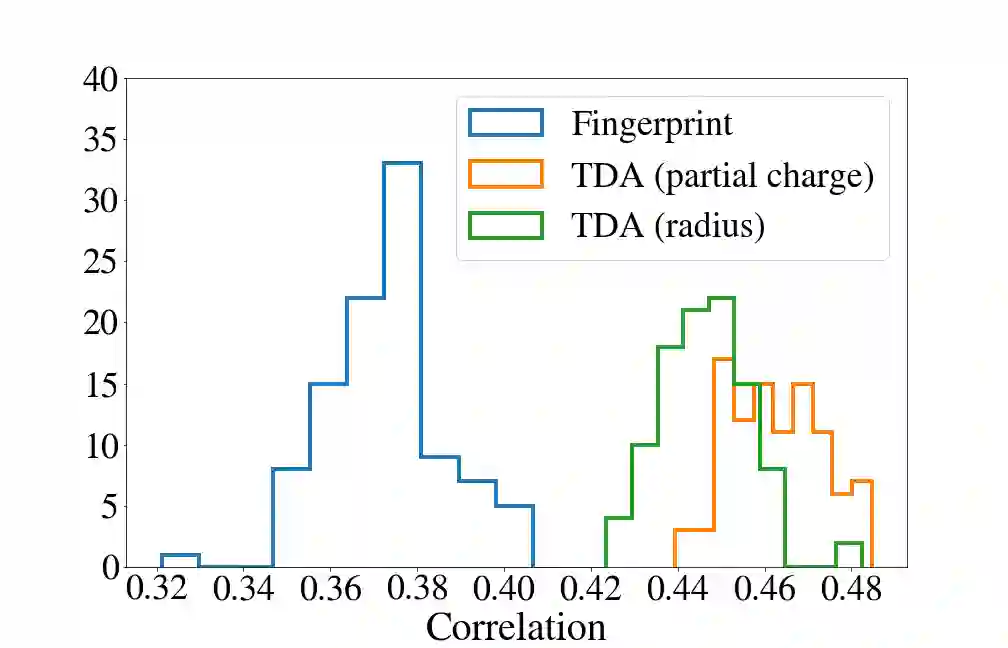
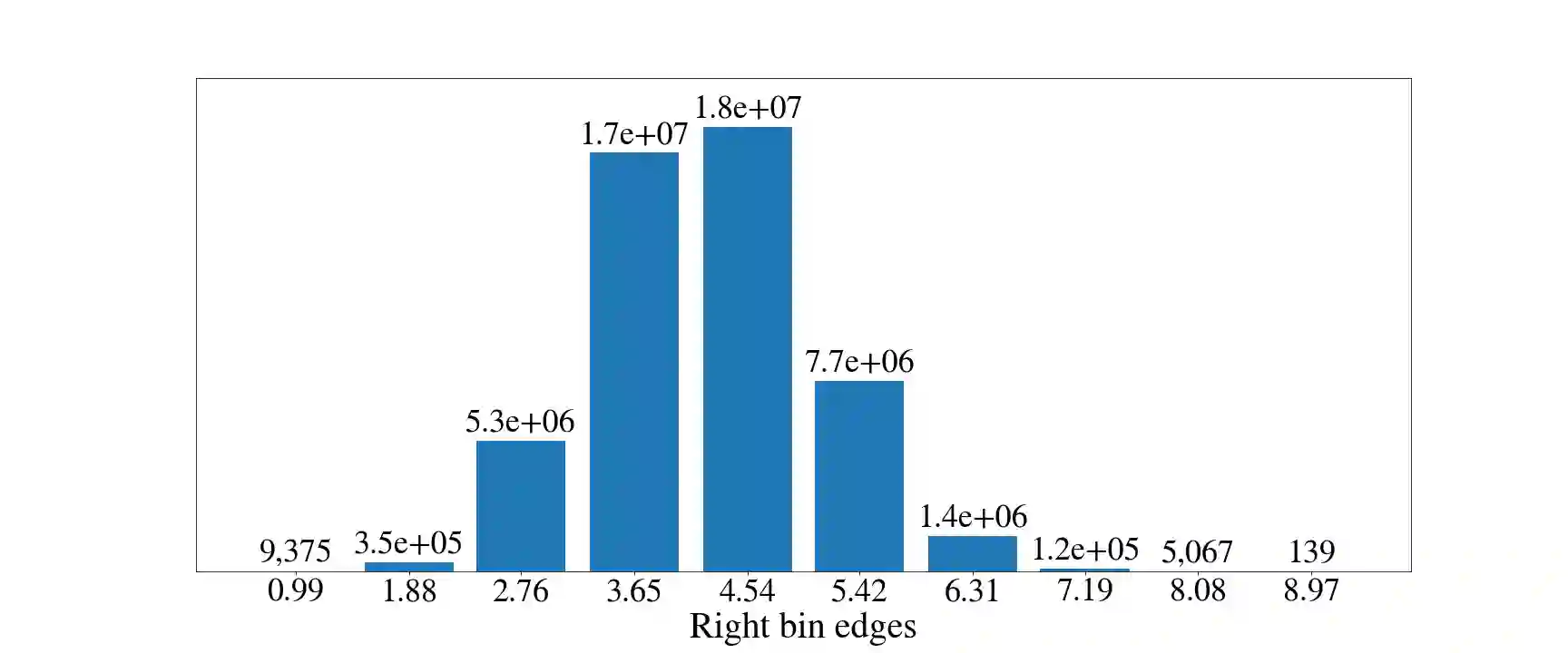
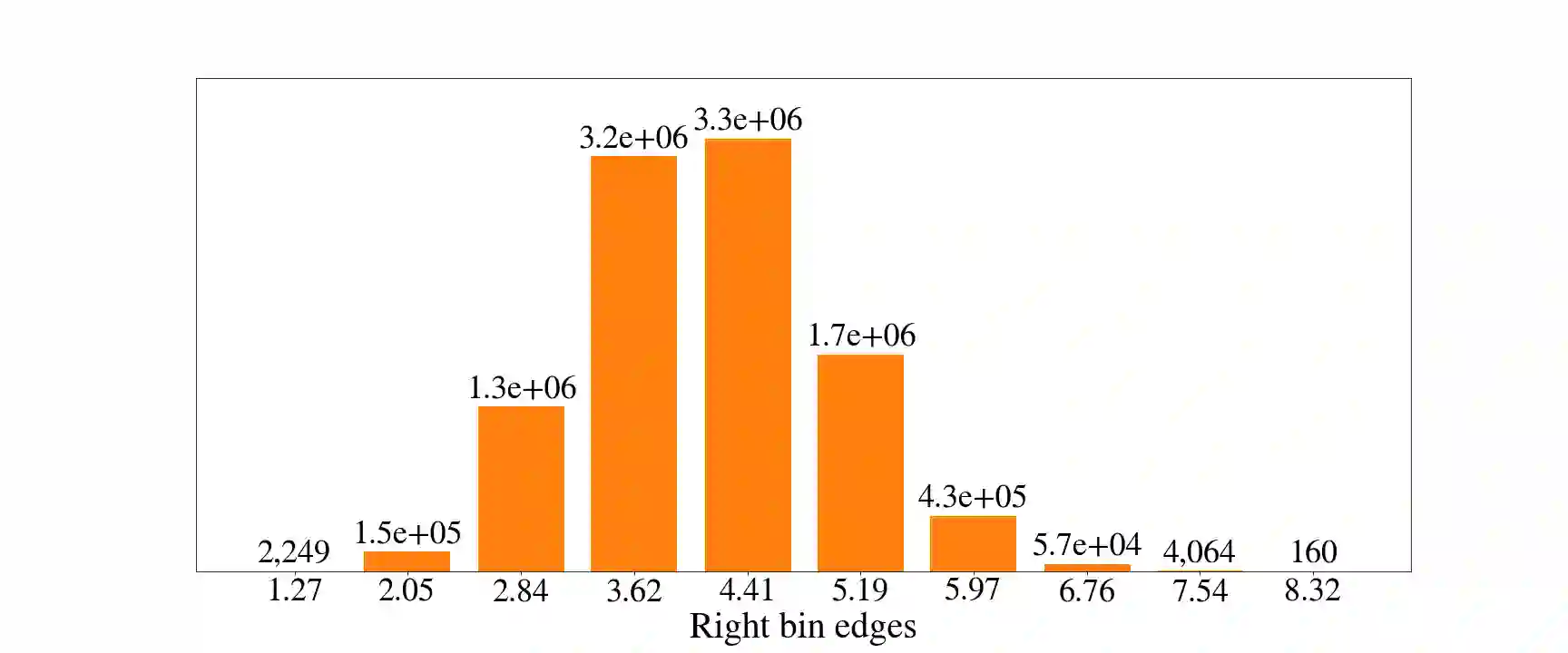
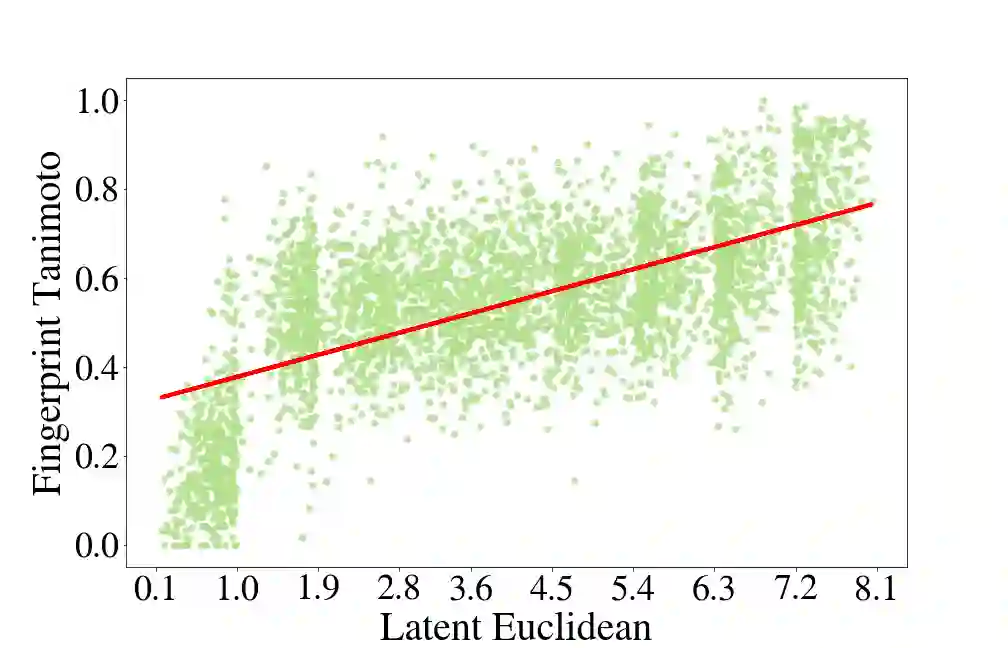
Deep generative models are increasingly becoming integral parts of the in silico molecule design pipeline and have dual goals of learning the chemical and structural features that render candidate molecules viable while also being flexible enough to generate novel designs. Specifically, Variational Auto Encoders (VAEs) are generative models in which encoder-decoder network pairs are trained to reconstruct training data distributions in such a way that the latent space of the encoder network is smooth. Therefore, novel candidates can be found by sampling from this latent space. However, the scope of architectures and hyperparameters is vast and choosing the best combination for in silico discovery has important implications for downstream success. Therefore, it is important to develop a principled methodology for distinguishing how well a given generative model is able to learn salient molecular features. In this work, we propose a method for measuring how well the latent space of deep generative models is able to encode structural and chemical features of molecular datasets by correlating latent space metrics with metrics from the field of topological data analysis (TDA). We apply our evaluation methodology to a VAE trained on SMILES strings and show that 3D topology information is consistently encoded throughout the latent space of the model.
翻译:深层基因模型正在日益成为硅分子设计管道中不可分割的组成部分,并且具有双重目标,即学习使候选分子具有可行性的化学和结构特征,同时又具有足够的灵活性,以产生新设计。具体地说,变形自动编码器(VAE)是基因模型,在这种模型中,对编码器脱coder网络网配对进行训练,以重建培训数据分布方式,使编码器网络的潜伏空间平滑。因此,可以通过从这一潜伏空间取样找到新的候选人。然而,建筑和超光度计的范围很广,选择硅发现的最佳组合对下游成功具有重要影响。因此,必须制定原则性方法,以区分给定的基因模型能够学习显著分子特征的好坏程度。在这项工作中,我们提出了一个方法,用以测量深层基因模型的潜在空间空间空间数据集的构造和化学特性,将潜在空间指标与表层数据分析领域的指标联系起来(TDA)。我们把评估方法应用到一个连续的VAE 3号模型,在SMILES上持续地展示了SMILES的高级模型。