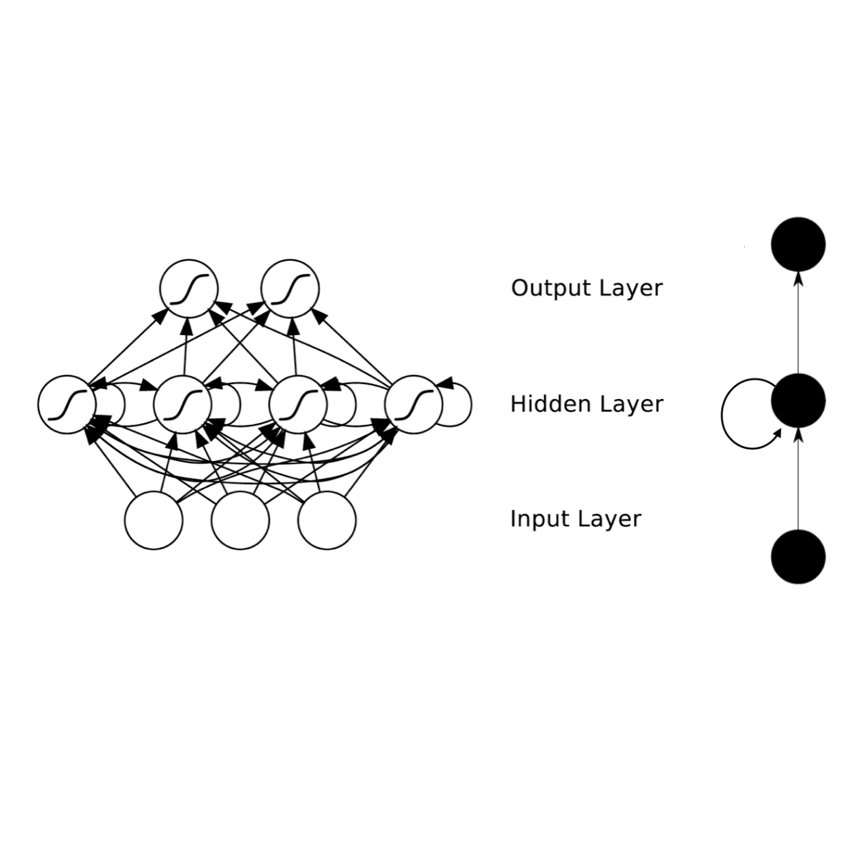
Methods for time series prediction and classification of gene regulatory networks (GRNs) from gene expression data have been treated separately so far. The recent emergence of attention-based recurrent neural networks (RNN) models boosted the interpretability of RNN parameters, making them appealing for the understanding of gene interactions. In this work, we generated synthetic time series gene expression data from a range of archetypal GRNs and we relied on a dual attention RNN to predict the gene temporal dynamics. We show that the prediction is extremely accurate for GRNs with different architectures. Next, we focused on the attention mechanism of the RNN and, using tools from graph theory, we found that its graph properties allow to hierarchically distinguish different architectures of the GRN. We show that the GRNs respond differently to the addition of noise in the prediction by the RNN and we relate the noise response to the analysis of the attention mechanism. In conclusion, this work provides a a way to understand and exploit the attention mechanism of RNN and it paves the way to RNN-based methods for time series prediction and inference of GRNs from gene expression data.
翻译:基因管理网络的时间序列预测和分类方法与基因表达数据迄今已分开处理。最近出现了关注的经常性神经网络模型,这提高了基因网络参数的可解释性,促使它们呼吁理解基因互动。在这项工作中,我们从一系列古老的基因网络中生成了合成的时间序列基因表达数据,我们依赖基因管理网络的双重关注来预测基因时间动态。我们表明,预测对于具有不同结构的基因管理网络来说非常精确。接下来,我们侧重于注意网络的注意机制,并利用图表理论的工具,我们发现其图表特性可以按等级区分基因网络的结构。我们表明,基因网络在预测中增加了噪音,我们把噪音反应与关注机制的分析联系起来。最后,这项工作为理解和利用基因网络的注意机制提供了一种途径,并为基于时间序列的预测方法和基因表达数据的推断铺平了道路。