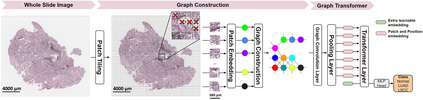
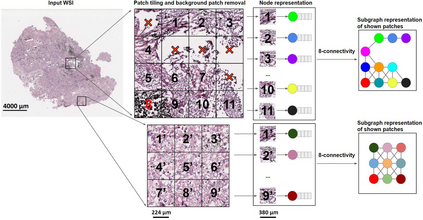
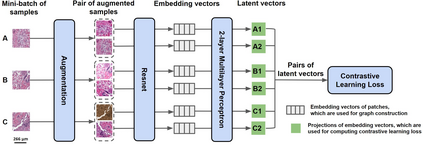
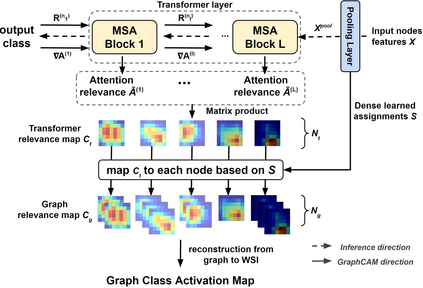
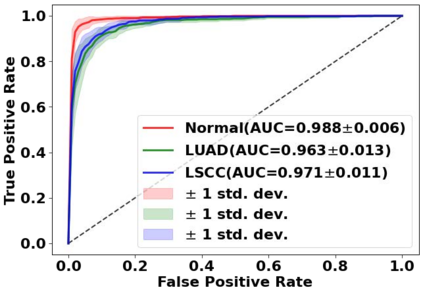
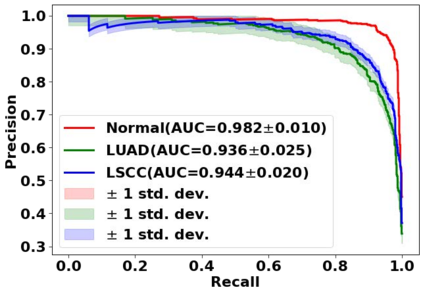
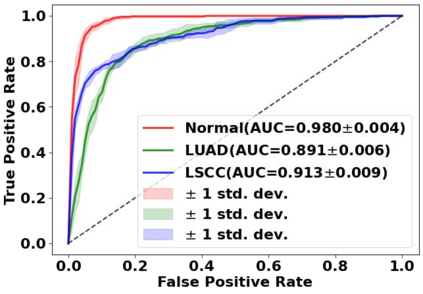
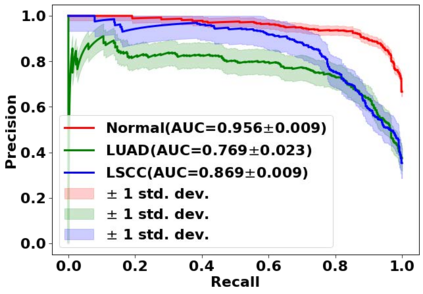
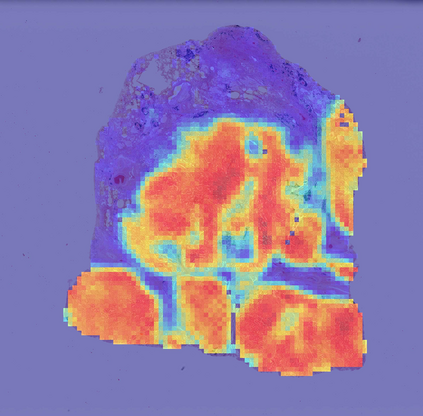
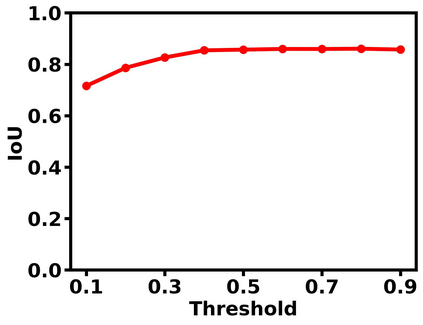
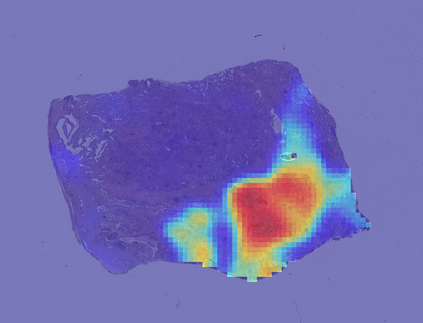
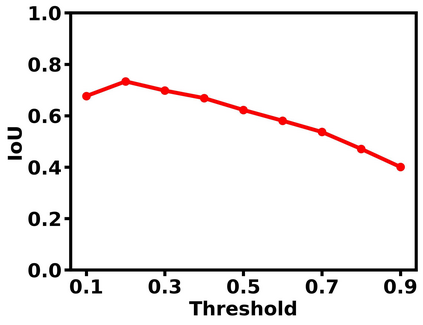
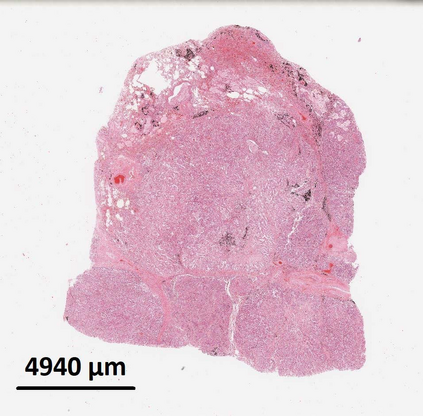
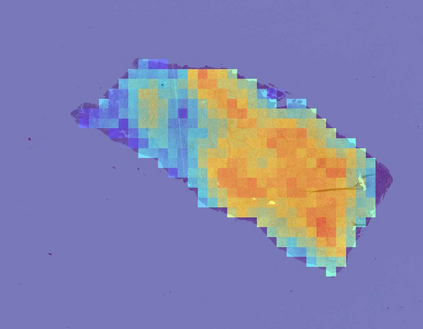
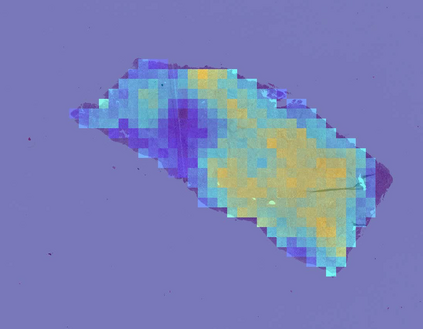
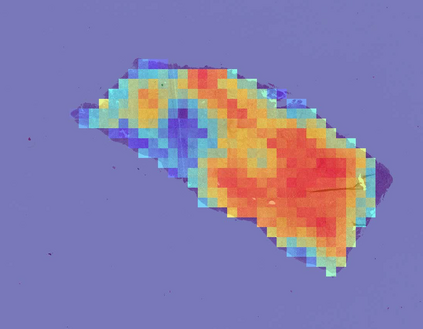
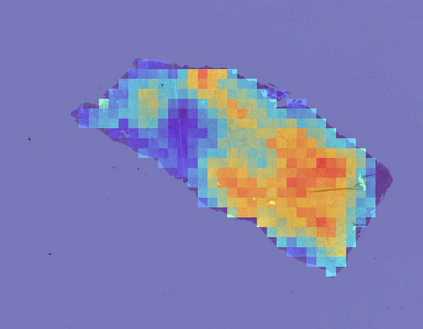
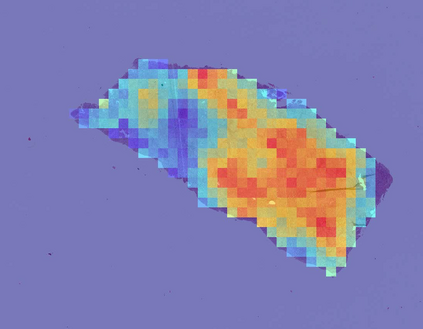
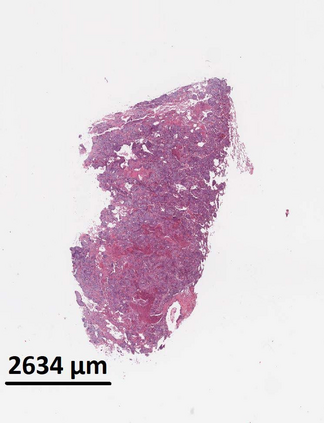
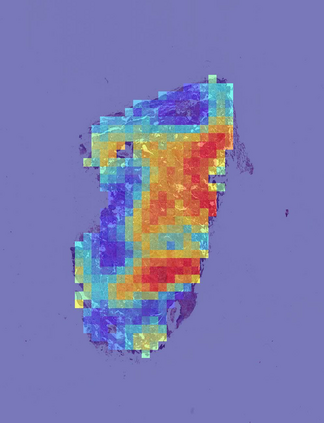
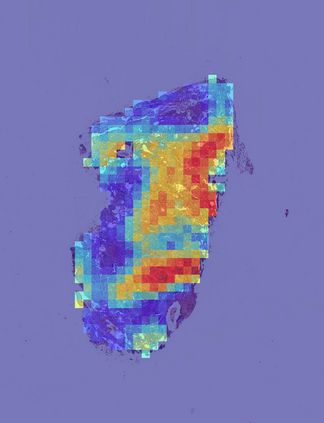
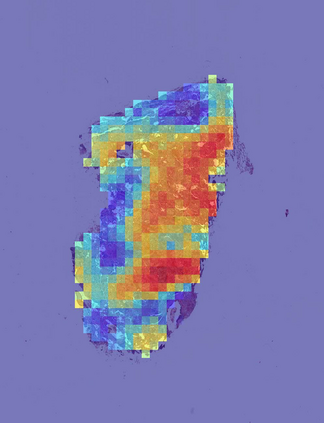
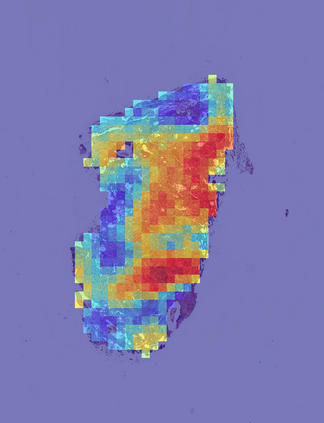
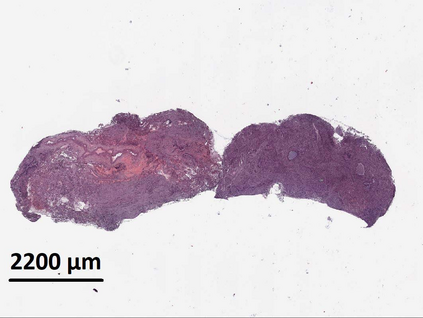
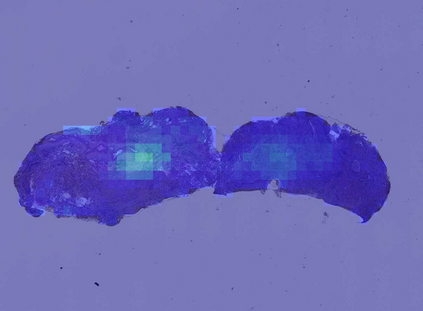
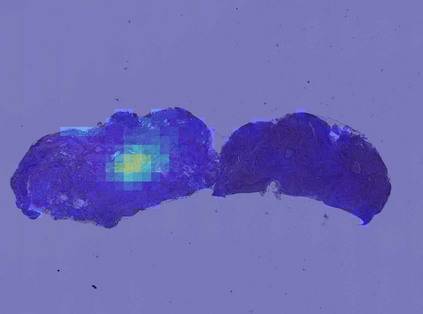
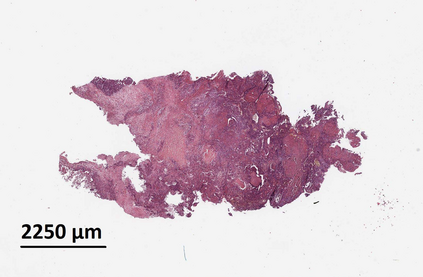
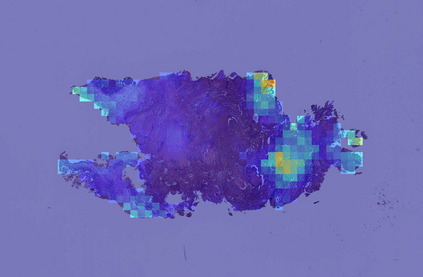
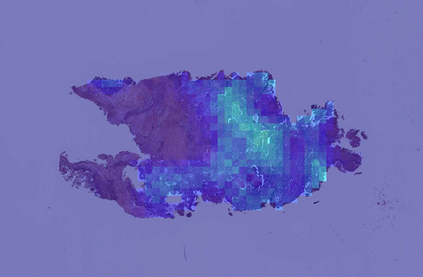
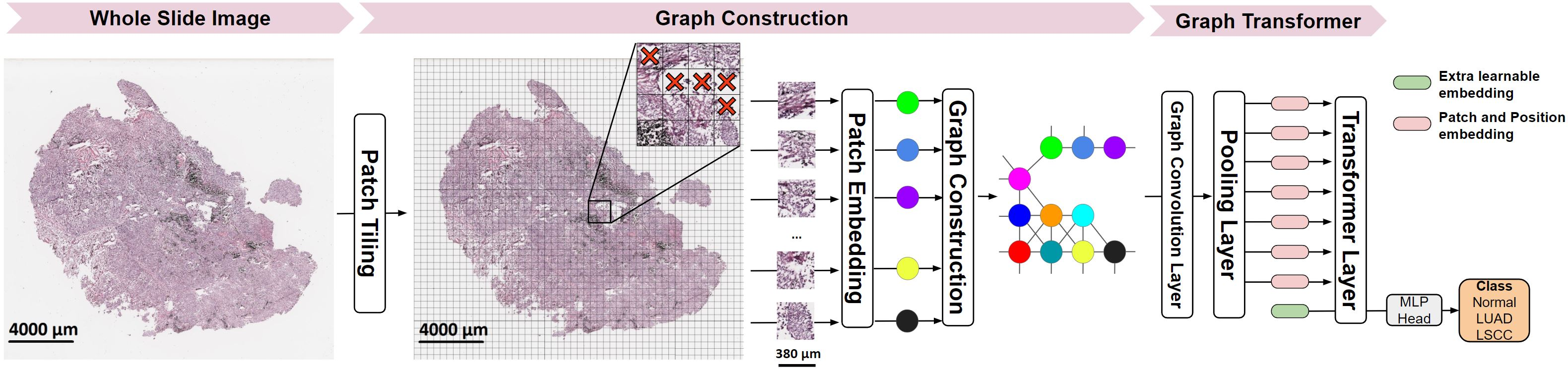
Deep learning is a powerful tool for whole slide image (WSI) analysis. Typically, when performing supervised deep learning, a WSI is divided into small patches, trained and the outcomes are aggregated to estimate disease grade. However, patch-based methods introduce label noise during training by assuming that each patch is independent with the same label as the WSI and neglect overall WSI-level information that is significant in disease grading. Here we present a Graph-Transformer (GT) that fuses a graph-based representation of an WSI and a vision transformer for processing pathology images, called GTP, to predict disease grade. We selected $4,818$ WSIs from the Clinical Proteomic Tumor Analysis Consortium (CPTAC), the National Lung Screening Trial (NLST), and The Cancer Genome Atlas (TCGA), and used GTP to distinguish adenocarcinoma (LUAD) and squamous cell carcinoma (LSCC) from adjacent non-cancerous tissue (normal). First, using NLST data, we developed a contrastive learning framework to generate a feature extractor. This allowed us to compute feature vectors of individual WSI patches, which were used to represent the nodes of the graph followed by construction of the GTP framework. Our model trained on the CPTAC data achieved consistently high performance on three-label classification (normal versus LUAD versus LSCC: mean accuracy$= 91.2$ $\pm$ $2.5\%$) based on five-fold cross-validation, and mean accuracy $= 82.3$ $\pm$ $1.0\%$ on external test data (TCGA). We also introduced a graph-based saliency mapping technique, called GraphCAM, that can identify regions that are highly associated with the class label. Our findings demonstrate GTP as an interpretable and effective deep learning framework for WSI-level classification.
翻译:深度学习是整个幻灯片图像( WSII) 分析的有力工具。 通常, 当进行有监督的深层次学习时, WSI 被分为小的补丁、 培训并汇总结果以估计疾病等级。 但是, 以补丁为基础的方法在培训过程中引入标签噪音, 假设每个补丁与 WSI 标签是独立的, 忽视了在疾病分级中具有重大意义的整体 WSI 级信息。 这里我们展示了一个图形- Transferent (GT), 将一个基于图形的 WSI 代表和一个用于处理病情图图像的视觉变异器, 称为 GTP 。 我们从临床蛋白质分析联合会( CPTAC ) 、 全国肺部筛选试验(NLST) 和癌症基因基因基因组(TC GGGGGGGA) 系统, 并使用GTP 高蛋白色素( LUAT) 和 高价值组织( 常规组织) 数据框架, 我们开发了一个对比学习框架, 也就是了我们所学的Oral- dalalalalalalalalal- dald 数据 。 这让我们用了C 数据框架, 用了C ASaltial- cald 数据 。