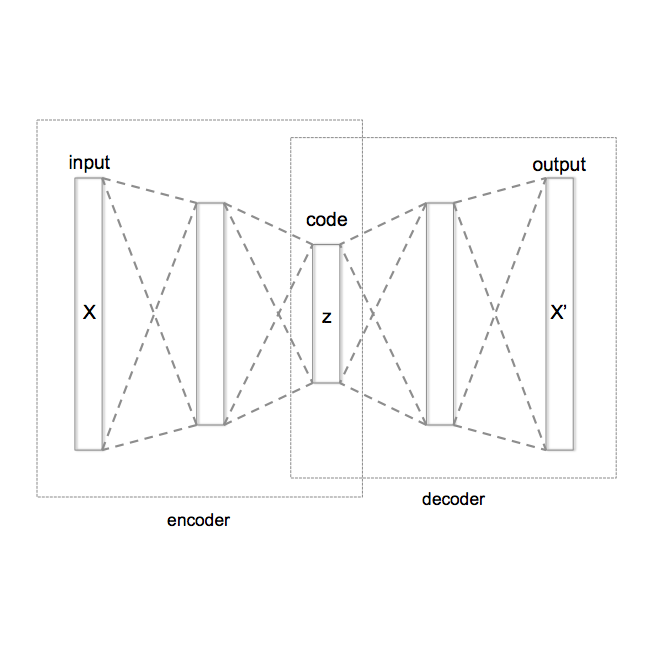
Deep Learning (DL) methods have dramatically increased in popularity in recent years. While its initial success was demonstrated in the classification and manipulation of image data, there has been significant growth in the application of DL methods to problems in the biomedical sciences. However, the greater prevalence and complexity of missing data in biomedical datasets present significant challenges for DL methods. Here, we provide a formal treatment of missing data in the context of Variational Autoencoders (VAEs), a popular unsupervised DL architecture commonly utilized for dimension reduction, imputation, and learning latent representations of complex data. We propose a new VAE architecture, NIMIWAE, that is one of the first to flexibly account for both ignorable and non-ignorable patterns of missingness in input features at training time. Following training, samples can be drawn from the approximate posterior distribution of the missing data can be used for multiple imputation, facilitating downstream analyses on high dimensional incomplete datasets. We demonstrate through statistical simulation that our method outperforms existing approaches for unsupervised learning tasks and imputation accuracy. We conclude with a case study of an EHR dataset pertaining to 12,000 ICU patients containing a large number of diagnostic measurements and clinical outcomes, where many features are only partially observed.
翻译:近些年来,深入学习(DL)方法的普及程度急剧提高。虽然在图像数据的分类和操作方面显示了最初的成功,但在应用DL方法处理生物医学科学问题方面却取得了显著的成绩;然而,生物医学数据集中缺失的数据更加普遍和复杂,给DL方法带来了重大挑战。在这里,我们正式处理在变形自动编码器(VAE)中缺失的数据,这是一个流行的、不受监督的DL结构,通常用于减少尺寸、估算和学习复杂数据的潜在表现。我们提出了一个新的VAE结构,NIMIWAE,这是首次灵活地说明在培训时间输入特征中可忽略和不可忽略的模式之一。在培训之后,可以从缺失数据的近似表面分布中抽取样本,用于多发,便利对高维度不完整数据集进行下游分析。我们通过统计模拟表明,我们的方法超越了现有的不协调的学习任务和倾斜度精确度的精确度。我们通过案例研究,得出了大部分EHR的临床测量结果,其中只有EHR的诊断结果。