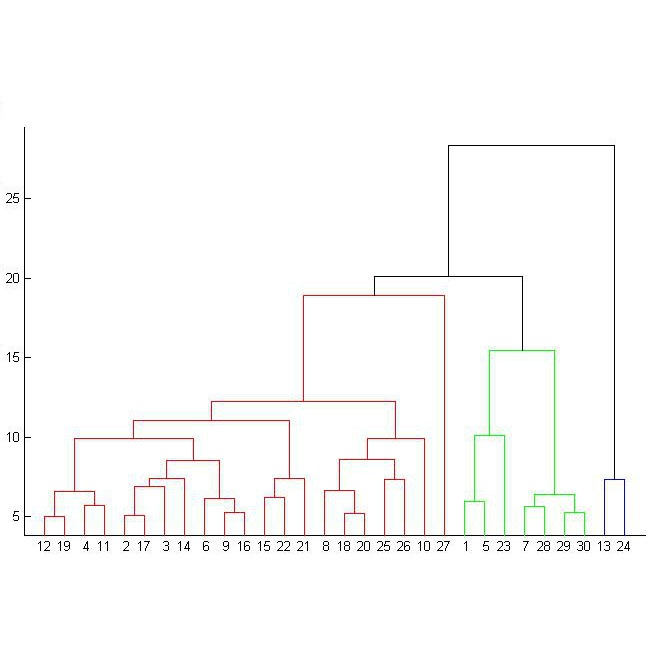
Datasets in the real world are often complex and to some degree hierarchical, with groups and sub-groups of data sharing common characteristics at different levels of abstraction. Understanding and uncovering the hidden structure of these datasets is an important task that has many practical applications. To address this challenge, we present a new and general method for building relational data trees by exploiting the learning dynamics of the Restricted Boltzmann Machine (RBM). Our method is based on the mean-field approach, derived from the Plefka expansion, and developed in the context of disordered systems. It is designed to be easily interpretable. We tested our method in an artificially created hierarchical dataset and on three different real-world datasets (images of digits, mutations in the human genome, and a homologous family of proteins). The method is able to automatically identify the hierarchical structure of the data. This could be useful in the study of homologous protein sequences, where the relationships between proteins are critical for understanding their function and evolution.
翻译:现实世界中的数据集往往复杂,在某种程度上等级分化,数据组和分组在不同抽象层次上共享共同特征。了解和发现这些数据集的隐藏结构是一项重要任务,具有许多实际应用。为了应对这一挑战,我们提出了一个新的通用方法,通过利用受限制的波尔茨曼机器(RBM)的学习动态来构建关联数据树。我们的方法基于从Plefka扩展中衍生出来的、在混乱系统背景下开发的平均值方法。它的设计容易解释。我们用人工创建的等级数据集和三种不同的真实世界数据集(数字图像、人类基因突变和蛋白质同系)测试了我们的方法(数字模型、人类基因突变和同质组)。这种方法能够自动识别数据的等级结构。这在对同质蛋白序列的研究中可能有用,因为同质蛋白质之间的关系对于了解其功能和演变至关重要。