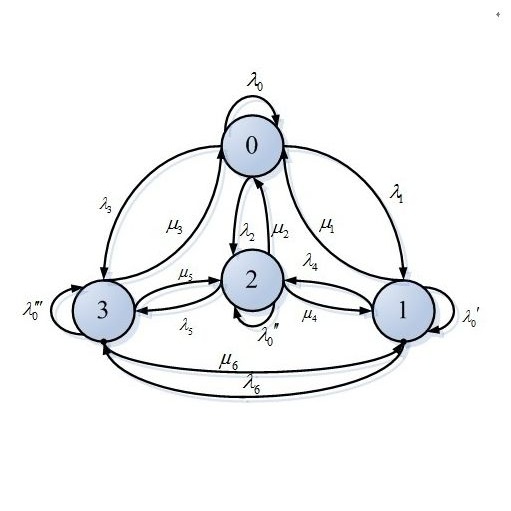
Stochastic kinetic models (SKMs) are increasingly used to account for the inherent stochasticity exhibited by interacting populations of species in areas such as epidemiology, population ecology and systems biology. Species numbers are modelled using a continuous-time stochastic process, and, depending on the application area of interest, this will typically take the form of a Markov jump process or an It\^o diffusion process. Widespread use of these models is typically precluded by their computational complexity. In particular, performing exact fully Bayesian inference in either modelling framework is challenging due to the intractability of the observed data likelihood, necessitating the use of computationally intensive techniques such as particle Markov chain Monte Carlo (particle MCMC). It is proposed to increase the computational and statistical efficiency of this approach by leveraging the tractability of an inexpensive surrogate derived directly from either the jump or diffusion process. The surrogate is used in three ways: in the design of a gradient-based parameter proposal, to construct an appropriate bridge and in the first stage of a delayed-acceptance step. The resulting approach, which exactly targets the posterior of interest, offers substantial gains in efficiency over a standard particle MCMC implementation.
翻译:随机动力学模型(SKMs)越来越被用于解释交互种群所表现出的内在随机性,其应用领域包括流行病学、种群生态学和系统生物学等。该模型使用连续时间随机过程来模拟物种数量,具体形式取决于所研究的应用领域,通常为马尔科夫跳跃过程或It\^ o扩散过程。由于观测数据似然函数的难以处理性,利用这些模型进行精确的贝叶斯推断是具有挑战性的,因此需要使用计算密集型的技术,如粒子马尔可夫链蒙特卡罗(particle MCMC)。本文提出了一种利用直接从跳跃或扩散过程中导出的廉价替代模型来增加该方法的计算和统计效率的方法。替代模型用于三个方面:在设计基于梯度的参数提议时,构建适当的桥梁,以及在延迟接受步骤的第一阶段中直接作为先验分布。所得到的方法可以精确地针对感兴趣的后验分布进行推断,并且相比于标准的粒子马尔可夫链蒙特卡罗实现具有显著的效率提升。