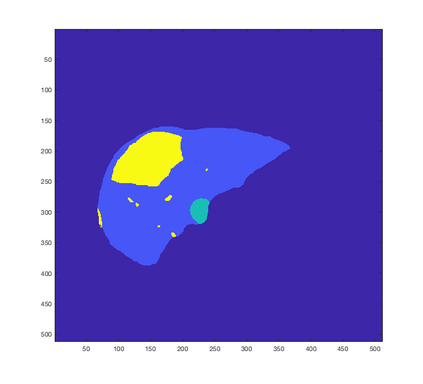
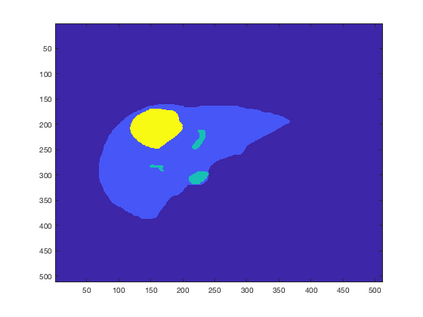
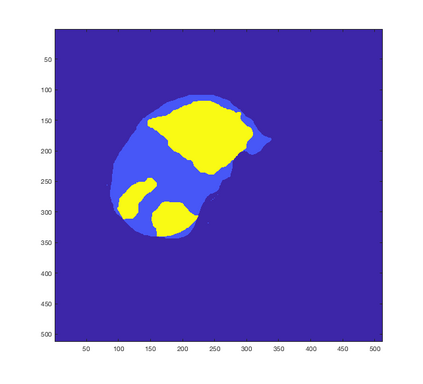
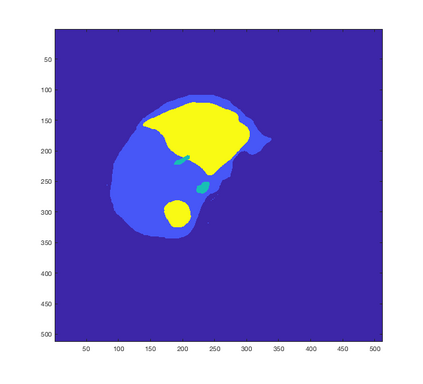
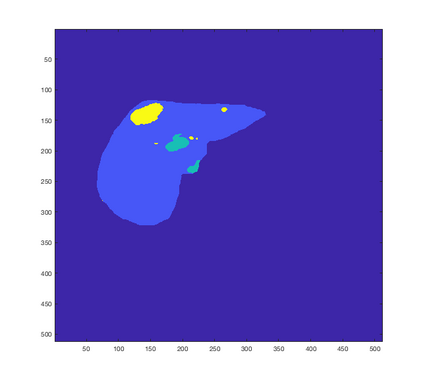
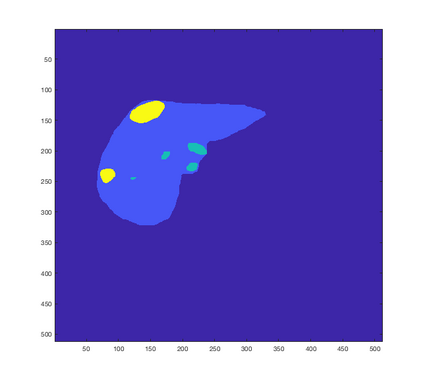
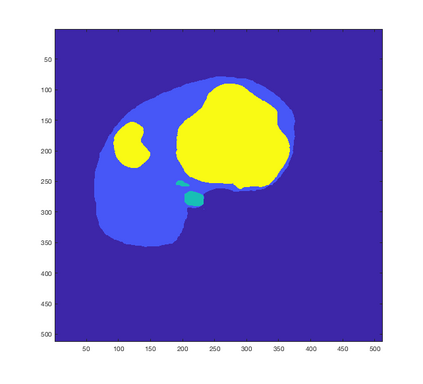
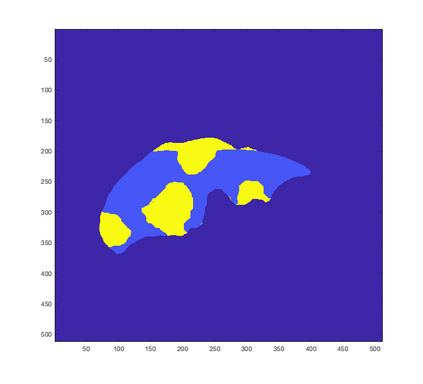
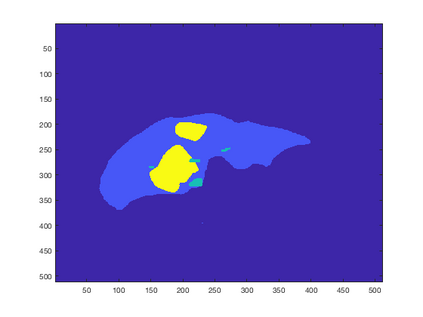
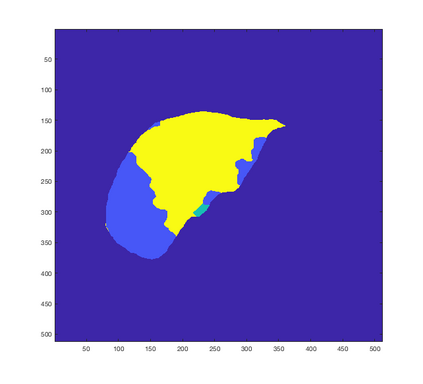
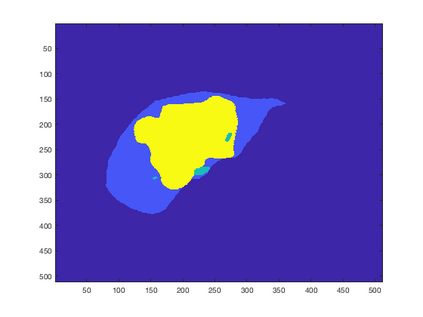
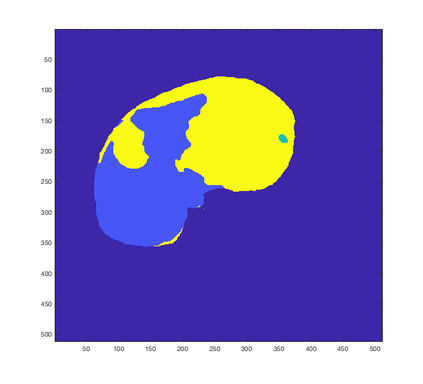
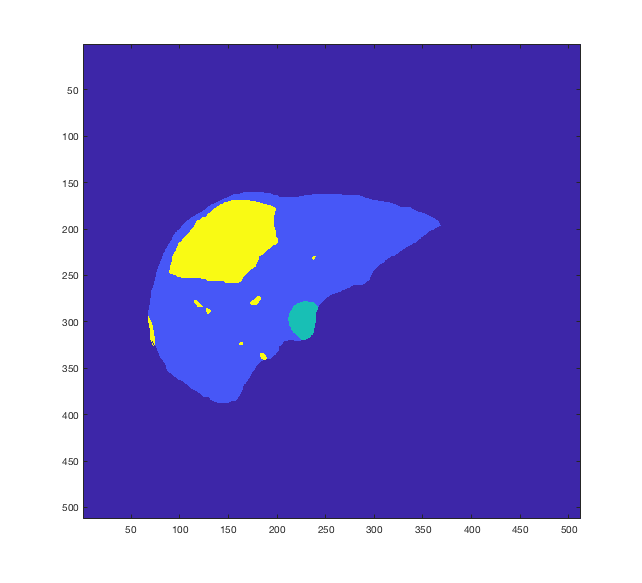
Tumor detection in biomedical imaging is a time-consuming process for medical professionals and is not without errors. Thus in recent decades, researchers have developed algorithmic techniques for image processing using a wide variety of mathematical methods, such as statistical modeling, variational techniques, and machine learning. In this paper, we propose a semi-automatic method for liver segmentation of 2D CT scans into three labels denoting healthy, vessel, or tumor tissue based on graph cuts. First, we create a feature vector for each pixel in a novel way that consists of the 59 intensity values in the time series data and propose a simplified perimeter cost term in the energy functional. We normalize the data and perimeter terms in the functional to expedite the graph cut without having to optimize the scaling parameter $\lambda$. In place of a training process, predetermined tissue means are computed based on sample regions identified by expert radiologists. The proposed method also has the advantage of being relatively simple to implement computationally. It was evaluated against the ground truth on a clinical CT dataset of 10 tumors and yielded segmentations with a mean Dice similarity coefficient (DSC) of .77 and mean volume overlap error (VOE) of 36.7%. The average processing time was 1.25 minutes per slice.
翻译:生物医学成像中的肿瘤检测对医疗专业人员来说是一个耗时的过程,而且并非没有错误。因此,近几十年来,研究人员利用多种数学方法,例如统计模型、变异技术和机器学习,开发了图像处理的算法技术。在本文中,我们建议采用半自动方法,将2DCT扫描分解成三个标签,以图解健康、容器或肿瘤组织。首先,我们以由时间序列数据59个强度值组成的新颖方式为每个像素创建了一个特性矢量,并提议在能源功能方面采用简化的周边成本术语。我们把功能中的数据和周边术语标准化,以加快图形的切除速度,而不必优化缩放参数 $\lambda$。在培训过程中,根据专家放射学家确定的样本区域来计算预定的组织手段。提议的方法还具有相对简单的应用优势。根据10个临床CT数据集的地面事实进行了评估,并产生了与平均狄氏度系数(DSSC)平均时间值7.77和平均速度误差36E。