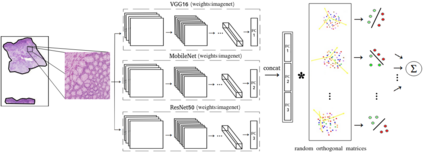
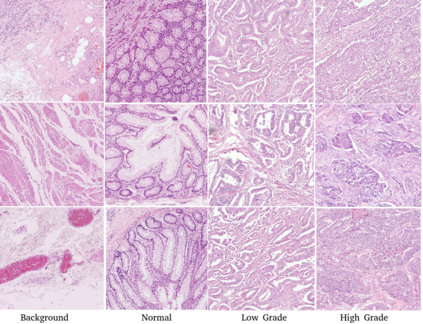
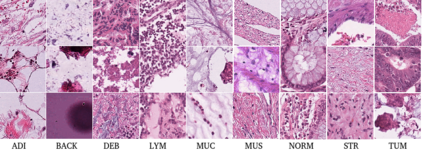
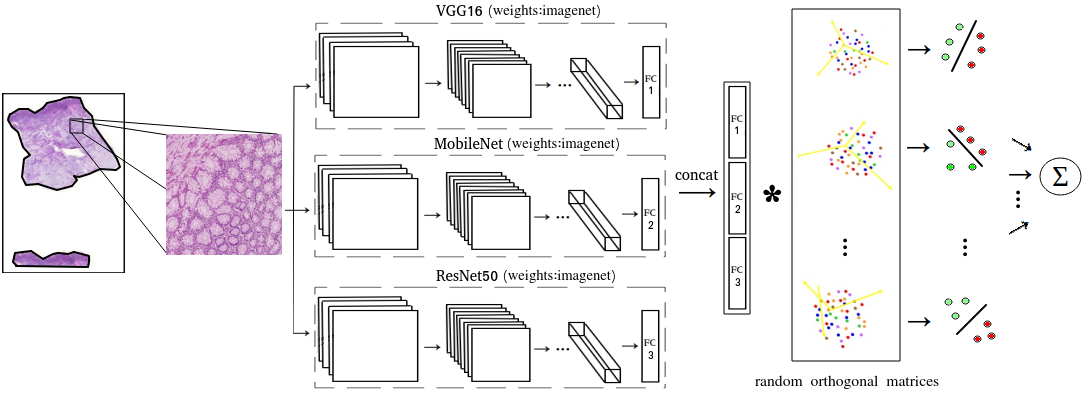
Computational pathology tasks have some unique characterises such as multi-gigapixel images, tedious and frequently uncertain annotations, and unavailability of large number of cases [13]. To address some of these issues, we present Deep Fastfood Ensembles - a simple, fast and yet effective method for combining deep features pooled from popular CNN models pre-trained on totally different source domains (e.g., natural image objects) and projected onto diverse dimensions using random projections, the so-called Fastfood [11]. The final ensemble output is obtained by a consensus of simple individual classifiers, each of which is trained on a different collection of random basis vectors. This offers extremely fast and yet effective solution, especially when training times and domain labels are of the essence. We demonstrate the effectiveness of the proposed deep fastfood ensemble learning as compared to the state-of-the-art methods for three different tasks in histopathology image analysis.
翻译:计算病理学任务有一些独特的特征,如多igapixel图像、乏味和经常不确定的附加说明以及大量案例[13]无法找到。为了解决其中的一些问题,我们介绍了深快食品集合(Deep Fastfood Enterfood Ansembles) -- -- 一种简单、快速且有效的方法,将广受欢迎的CNN模型的深层特征组合起来,在完全不同的源域(例如自然图像物体)上预先培训,并使用随机预测,即所谓的快食品[11],并投射到不同的维度上。最后的混合输出是通过简单的单个分类师的共识取得的,每个分类师都接受过不同随机矢量收集的培训。这提供了非常快速而有效的解决方案,特别是当培训时间和域标签是本质时。我们展示了拟议的深度快食品共习学习与他病理图像分析中三种不同任务的最新方法相比的有效性。