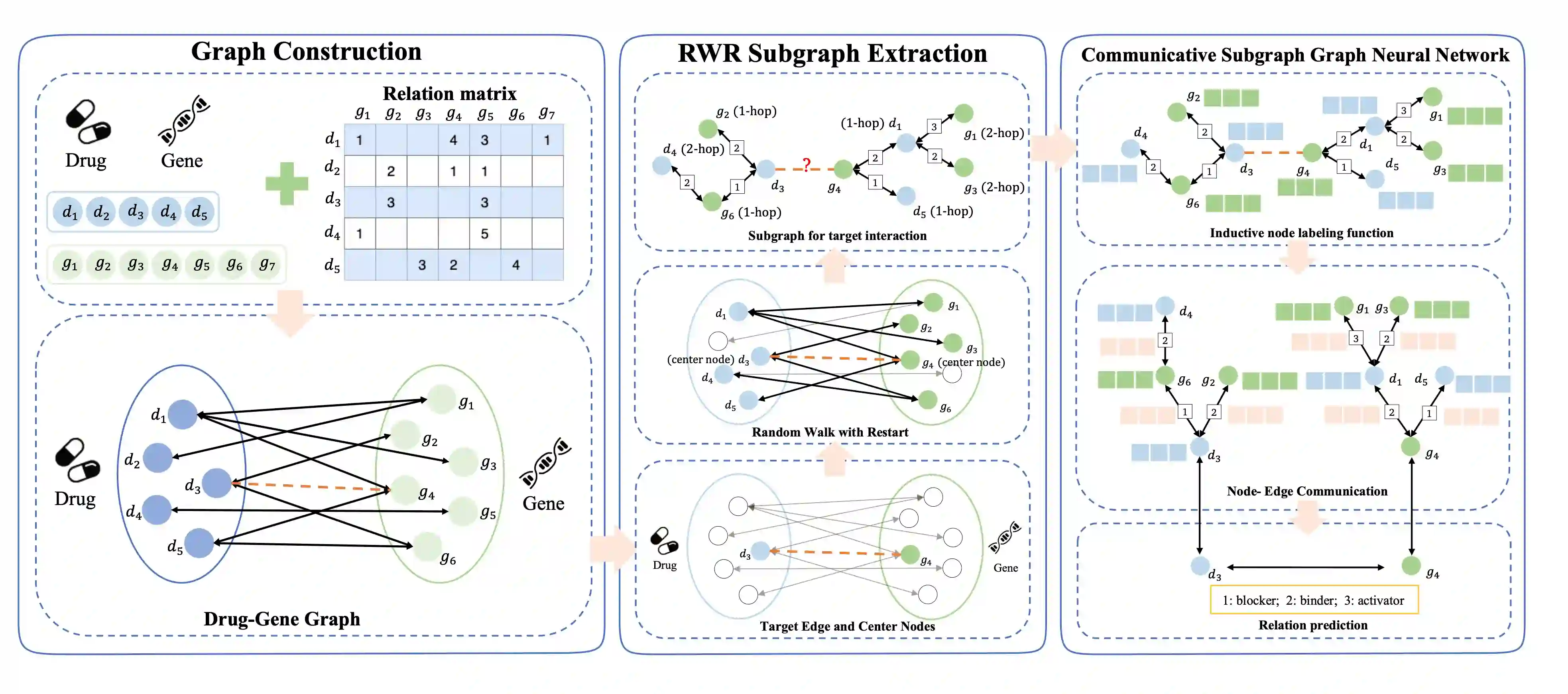
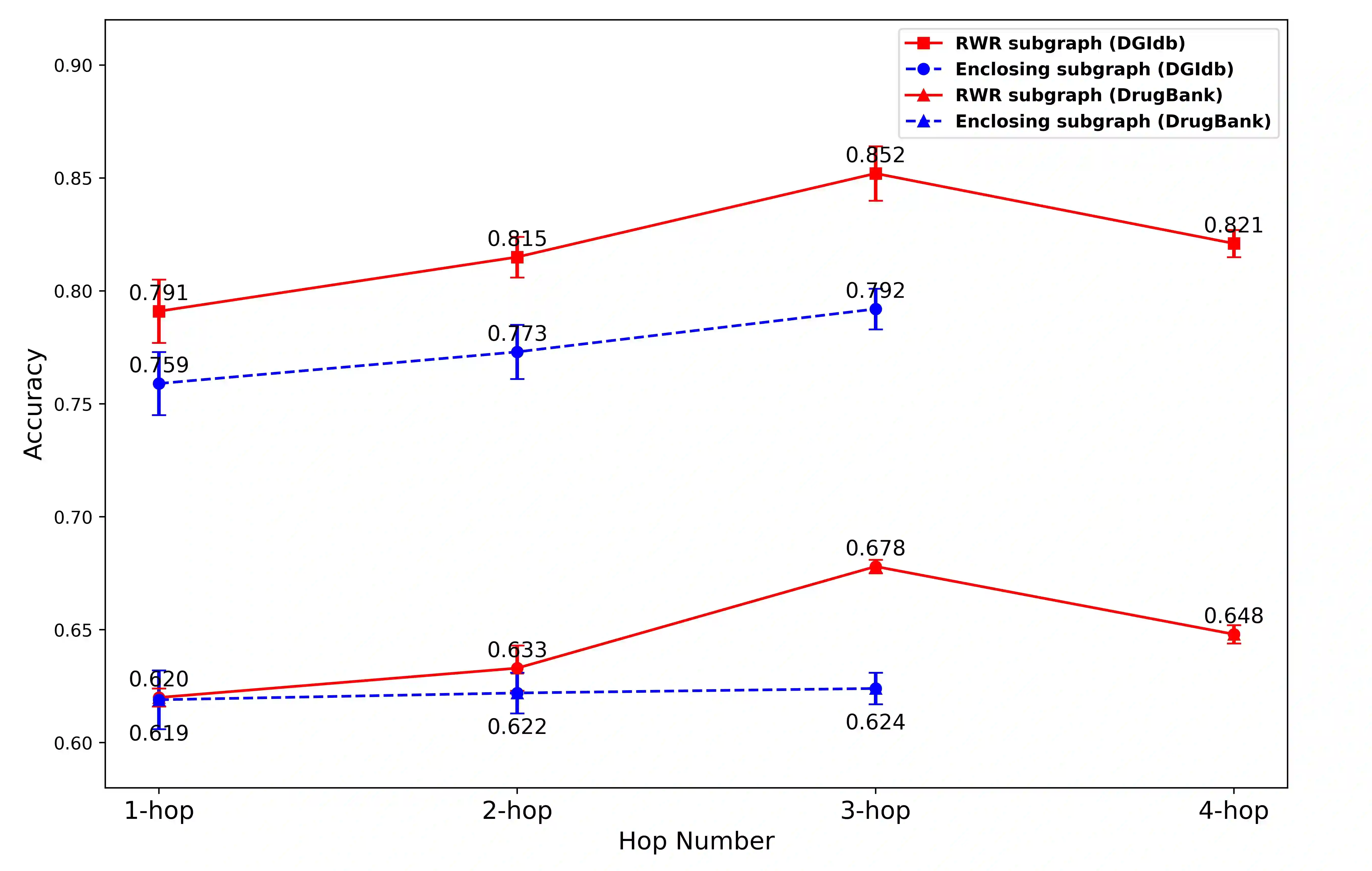
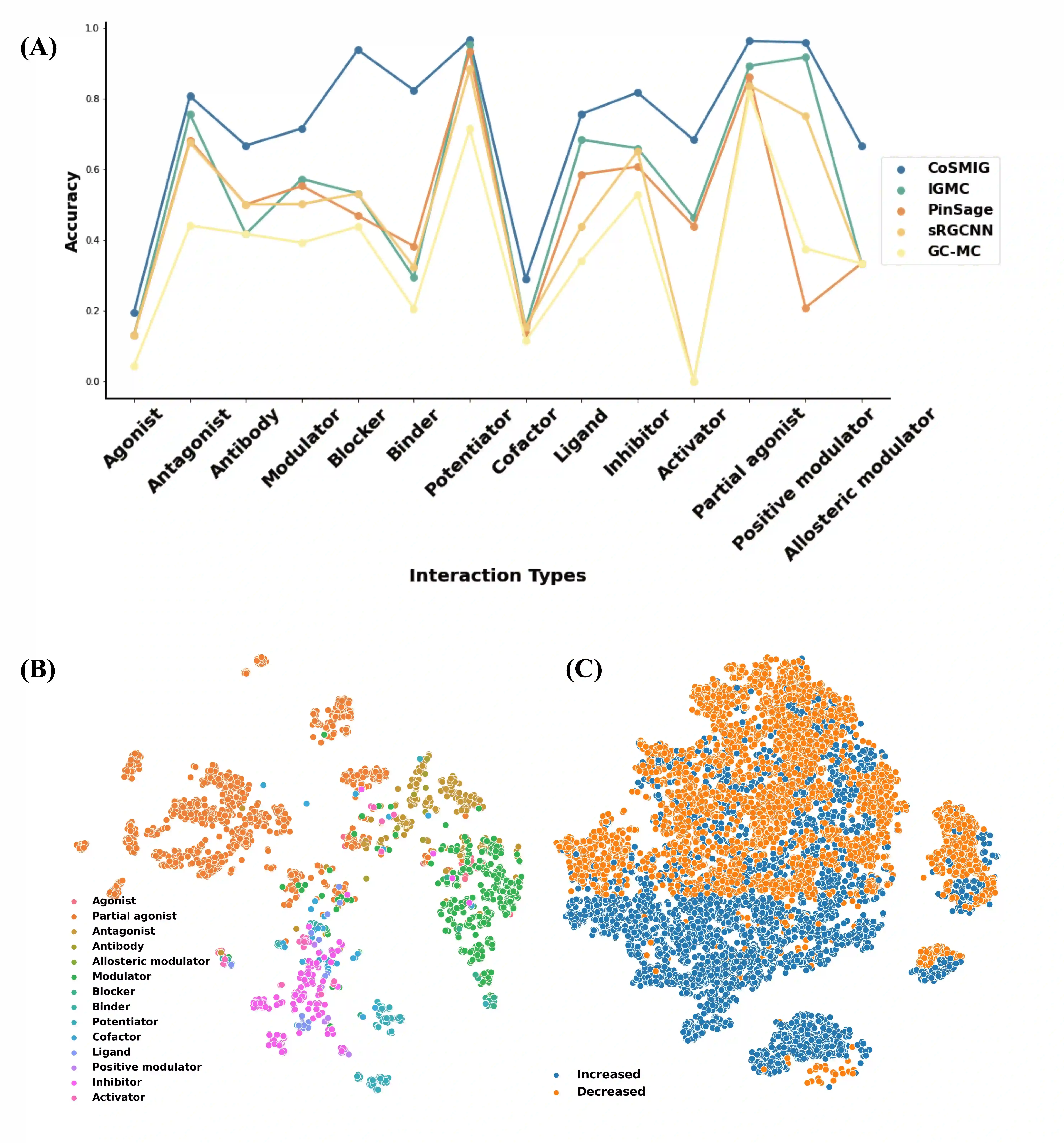
Illuminating the interconnections between drugs and genes is an important topic in drug development and precision medicine. Currently, computational predictions of drug-gene interactions mainly focus on the binding interactions without considering other relation types like agonist, antagonist, etc. In addition, existing methods either heavily rely on high-quality domain features or are intrinsically transductive, which limits the capacity of models to generalize to drugs/genes that lack external information or are unseen during the training process. To address these problems, we propose a novel Communicative Subgraph representation learning for Multi-relational Inductive drug-Gene interactions prediction (CoSMIG), where the predictions of drug-gene relations are made through subgraph patterns, and thus are naturally inductive for unseen drugs/genes without retraining or utilizing external domain features. Moreover, the model strengthened the relations on the drug-gene graph through a communicative message passing mechanism. To evaluate our method, we compiled two new benchmark datasets from DrugBank and DGIdb. The comprehensive experiments on the two datasets showed that our method outperformed state-of-the-art baselines in the transductive scenarios and achieved superior performance in the inductive ones. Further experimental analysis including LINCS experimental validation and literature verification also demonstrated the value of our model.
翻译:目前,药物基因相互作用的计算预测主要侧重于约束性互动,而没有考虑其他类型的关系,例如强力药剂、敌国药剂等。此外,现有的方法要么严重依赖高质量的领域特征,要么本质上是转基因性的,这就限制了模型向缺乏外部信息或培训过程中看不见的药物/基因进行概括的能力。为了解决这些问题,我们建议为多种关系诱导药物-基因相互作用预测(COSMIG)进行新型的交流分层代表学习。 在这两种数据组上,药物-基因关系预测是通过子图模式作出的,因此,在没有再培训或利用外部领域特征的情况下,自然地对看不见的药物/基因进行诱导。此外,该模型通过传播信息传递机制加强了药物-基因图上的关系。为了评估我们的方法,我们汇编了两个新的基准数据集,分别来自药物银行和DGIDb。 在两种数据组上进行的全面实验显示,我们的方法超越了在试算性基准中所展示的先进性能测试基准,其中包括在试算性实验性基准中所展示的先进性实验性基准。