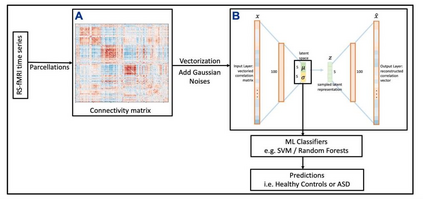
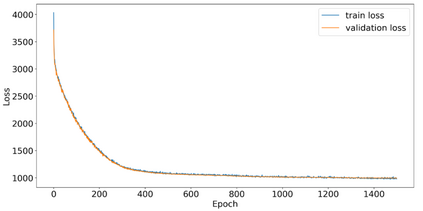
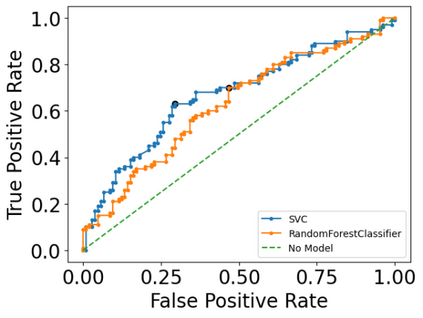
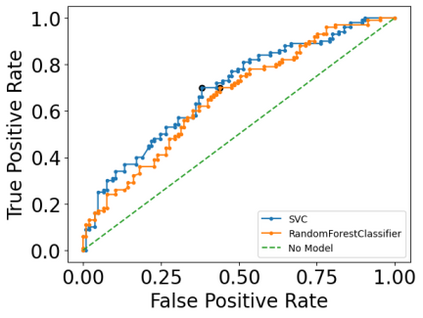
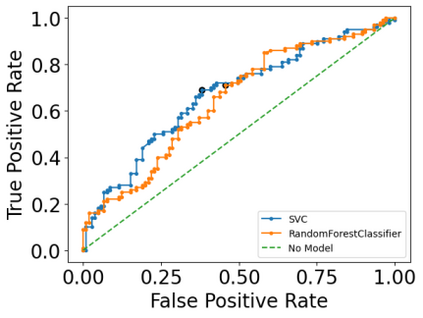
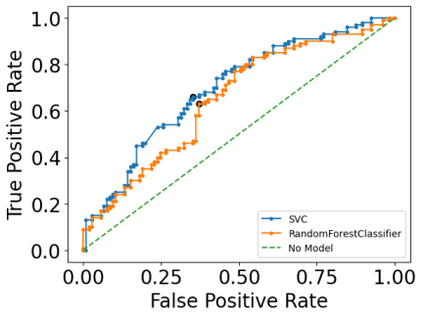
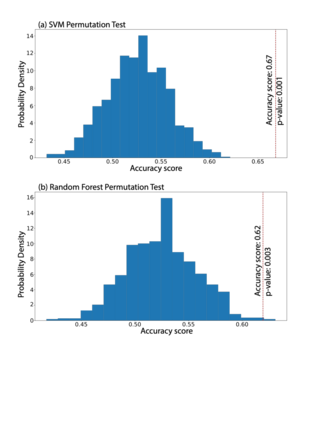
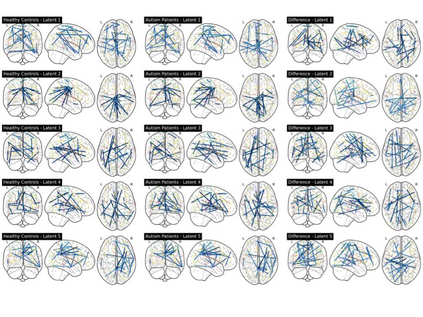
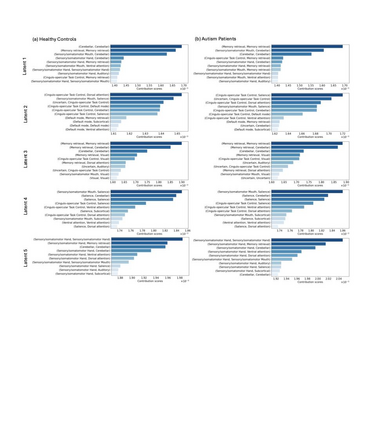
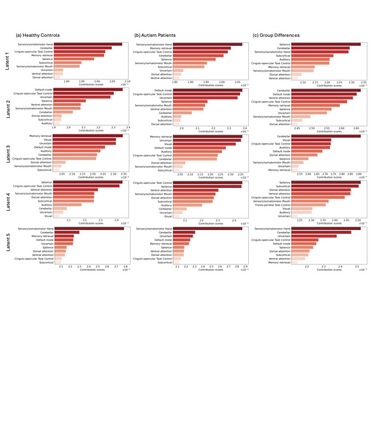
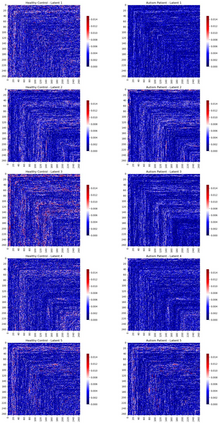
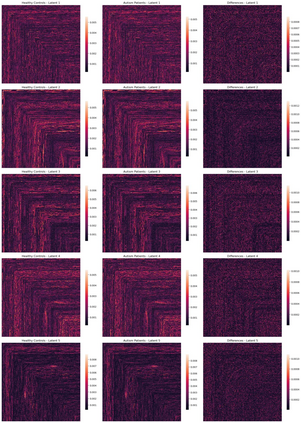
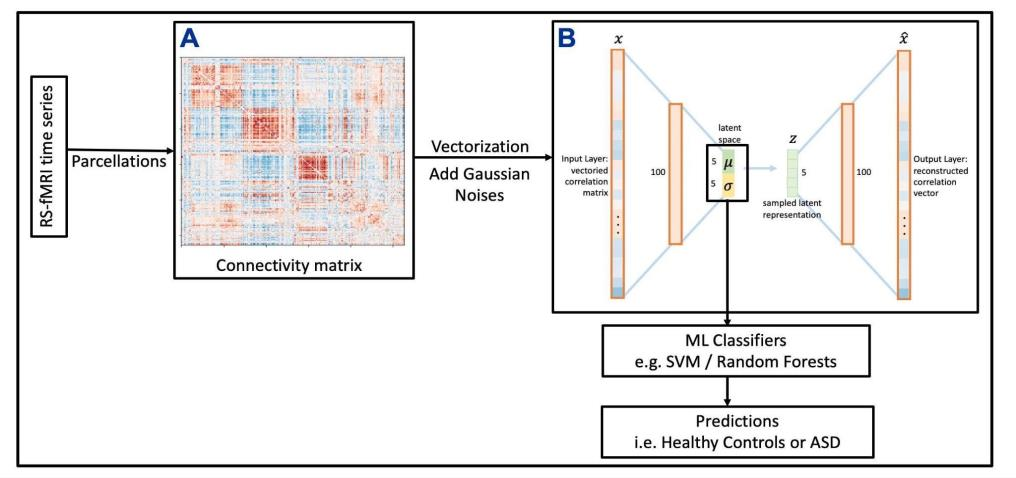
Autism spectrum disorders (ASDs) are developmental conditions characterized by restricted interests and difficulties in communication. The complexity of ASD has resulted in a deficiency of objective diagnostic biomarkers. Deep learning methods have gained recognition for addressing these challenges in neuroimaging analysis, but finding and interpreting such diagnostic biomarkers are still challenging computationally. Here, we propose a feature reduction pipeline using resting-state fMRI data. We used Craddock atlas and Power atlas to extract functional connectivity data from rs-fMRI, resulting in over 30 thousand features. By using a denoising variational autoencoder, our proposed pipeline further compresses the connectivity features into 5 latent Gaussian distributions, providing is a low-dimensional representation of the data to promote computational efficiency and interpretability. To test the method, we employed the extracted latent representations to classify ASD using traditional classifiers such as SVM on a large multi-site dataset. The 95% confidence interval for the prediction accuracy of SVM is [0.63, 0.76] after site harmonization using the extracted latent distributions. Without using DVAE for dimensionality reduction, the prediction accuracy is 0.70, which falls within the interval. The DVAE successfully encoded the diagnostic information from rs-fMRI data without sacrificing prediction performance. The runtime for training the DVAE and obtaining classification results from its extracted latent features was 7 times shorter compared to training classifiers directly on the raw data. Our findings suggest that the Power atlas provides more effective brain connectivity insights for diagnosing ASD than Craddock atlas. Additionally, we visualized the latent representations to gain insights into the brain networks contributing to the differences between ASD and neurotypical brains.
翻译:暂无翻译