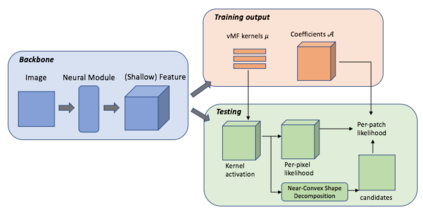
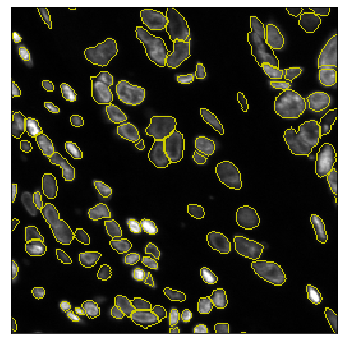
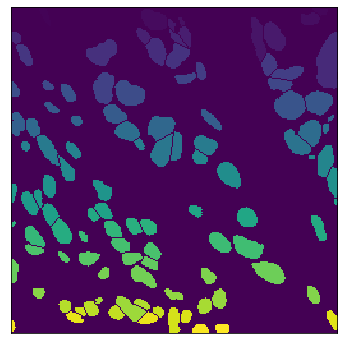
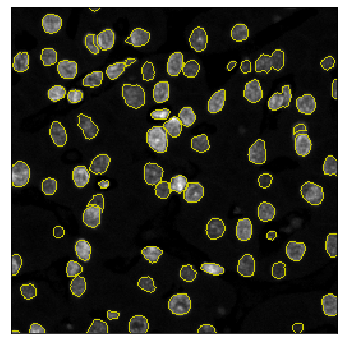
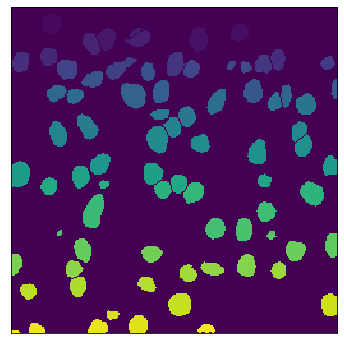
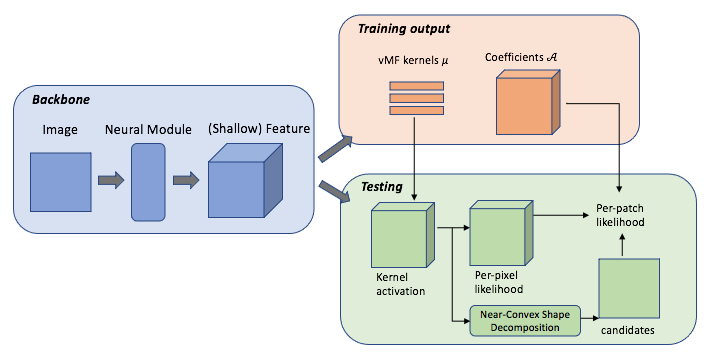
The field of computational pathology has witnessed great advancements since deep neural networks have been widely applied. These deep neural networks usually require large numbers of annotated data to train vast parameters. However, it takes significant effort to annotate a large histopathology dataset. We propose to build a data-efficient model, which only requires partial annotation, specifically on isolated nucleus, rather than on the whole slide image. It exploits shallow features as its backbone and is light-weight, therefore a small number of data is sufficient for training. What's more, it is a generative compositional model, which enjoys interpretability in its prediction. The proposed method could be an alternative solution for the data-hungry problem of deep learning methods.
翻译:自深神经网络广泛应用以来,计算病理学领域取得了巨大进步,这些深层神经网络通常需要大量附加说明的数据来训练庞大的参数,然而,它需要大量的努力来说明庞大的病理学数据集。我们建议建立一个数据效率模型,仅需要部分注解,特别是孤立核,而不是整个幻灯片图象。它利用浅浅的特征作为主干线,因此,少量数据足以用于培训。此外,它是一种基因化的构成模型,在预测中可加以解释。提议的方法可以作为深层学习方法的数据饥饿问题的替代解决办法。