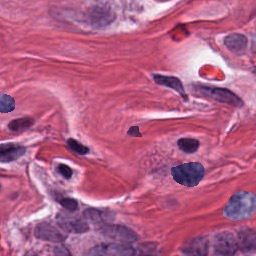
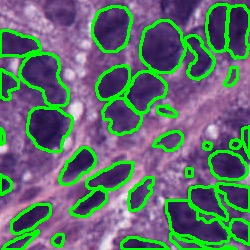
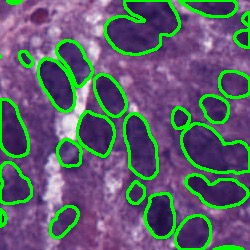
Nuclei Segmentation from histology images is a fundamental task in digital pathology analysis. However, deep-learning-based nuclei segmentation methods often suffer from limited annotations. This paper proposes a realistic data augmentation method for nuclei segmentation, named InsMix, that follows a Copy-Paste-Smooth principle and performs morphology-constrained generative instance augmentation. Specifically, we propose morphology constraints that enable the augmented images to acquire luxuriant information about nuclei while maintaining their morphology characteristics (e.g., geometry and location). To fully exploit the pixel redundancy of the background and improve the model's robustness, we further propose a background perturbation method, which randomly shuffles the background patches without disordering the original nuclei distribution. To achieve contextual consistency between original and template instances, a smooth-GAN is designed with a foreground similarity encoder (FSE) and a triplet loss. We validated the proposed method on two datasets, i.e., Kumar and CPS datasets. Experimental results demonstrate the effectiveness of each component and the superior performance achieved by our method to the state-of-the-art methods.
翻译:生理图象的内核分解是数字病理学分析的一项基本任务。然而,基于深学习的核分解方法往往受到有限的说明。本文件提出一个符合复制-帕斯特-松动原理的、符合复制-帕斯特-松动原理并进行形态-受限制的基因放大的切合实际的数据扩增方法InsMix。具体地说,我们提出形态限制,使放大的图象能够在保持其形态特征(例如几何和位置)的同时获得关于核的奢侈信息。为了充分利用背景的像素冗余并改进模型的坚固性,我们进一步提议一种背景透析方法,在不扰乱原始核分布的情况下随机地打乱背景补。为了在原始和模板之间实现背景一致性,我们设计了一个光色GAN,在保持其形态特征特性(例如几何和位置)的同时,能够获取关于核子的奢华信息。我们验证了两种数据集的拟议方法,即库马尔和CPS-PS-实验方法的效能,以我们每个部分的高级性能显示我们每个实验方法取得的结果。