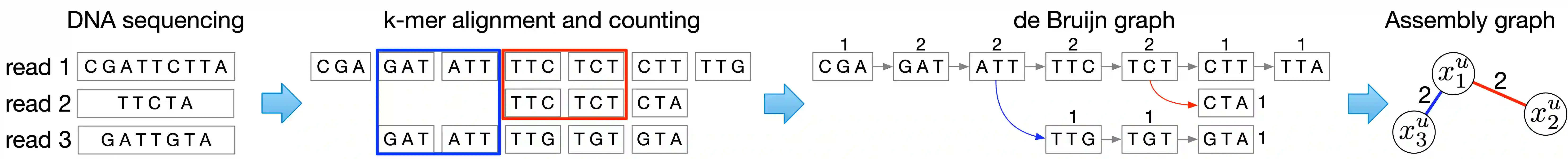
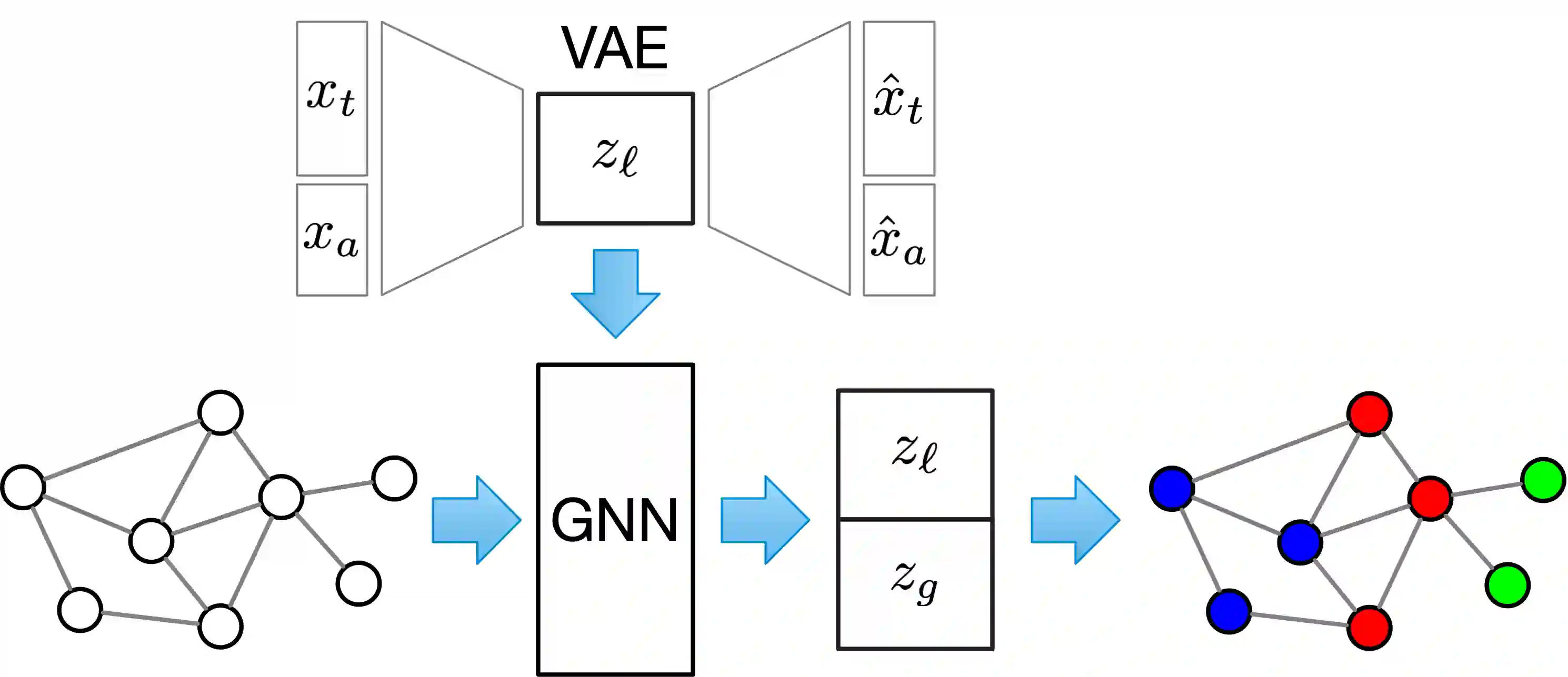
Microbes have a profound impact on our health and environment, but our understanding of the diversity and function of microbial communities is severely limited. Through DNA sequencing of microbial communities (metagenomics), DNA fragments (reads) of the individual microbes can be obtained, which through assembly graphs can be combined into long contiguous DNA sequences (contigs). Given the complexity of microbial communities, single contig microbial genomes are rarely obtained. Instead, contigs are eventually clustered into bins, with each bin ideally making up a full genome. This process is referred to as metagenomic binning. Current state-of-the-art techniques for metagenomic binning rely only on the local features for the individual contigs. These techniques therefore fail to exploit the similarities between contigs as encoded by the assembly graph, in which the contigs are organized. In this paper, we propose to use Graph Neural Networks (GNNs) to leverage the assembly graph when learning contig representations for metagenomic binning. Our method, VaeG-Bin, combines variational autoencoders for learning latent representations of the individual contigs, with GNNs for refining these representations by taking into account the neighborhood structure of the contigs in the assembly graph. We explore several types of GNNs and demonstrate that VaeG-Bin recovers more high-quality genomes than other state-of-the-art binners on both simulated and real-world datasets.
翻译:微生物对我们的健康和环境有着深远的影响,但我们对微生物群落多样性和功能的理解非常有限。 通过微生物群落(基因基因组)的DNA排序,可以获取单个微生物的DNA碎片(读数),通过组装图可以将其结合成长期毗连的DNA序列(连接)。鉴于微生物群落的复杂性,很少获得单一的同类微生物基因组。相反,我们建议最终将基因组分组成垃圾箱,每个垃圾箱最好能组成一个完整的基因组。这一过程被称为美化基因组。目前最先进的集成技术仅取决于单个基因组的本地特征。因此,这些技术无法利用组群群群群群群群群群群群群群群之间的相似之处。在本文件中,我们建议使用图形神经网(GNNNPs)来利用组群集的模拟图解算。这个过程被称为美化本组群集的模拟,目前最先进的集集技术仅取决于单个基因组群集的本地组群集结构。我们的方法,VaeG-B,这些基因组群集的模型模型将这些基因组群落的模型与GNG-NG-NF的模型的模型组合化图层模型组合化模型结合起来。