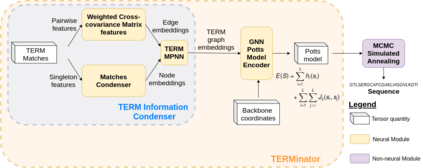
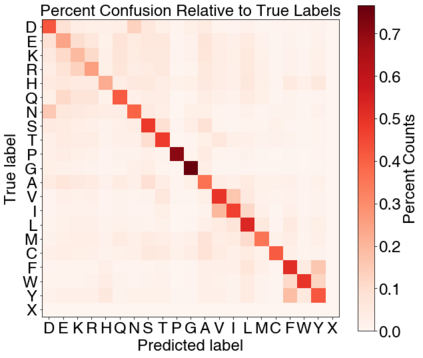
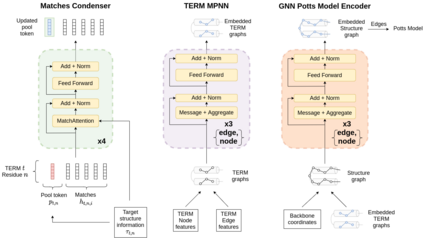
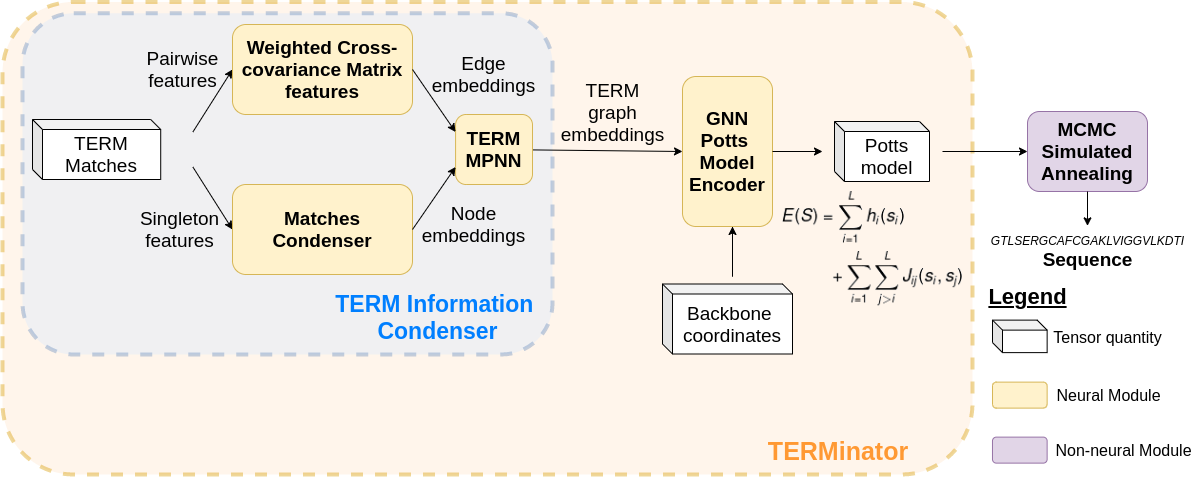
Computational protein design has the potential to deliver novel molecular structures, binders, and catalysts for myriad applications. Recent neural graph-based models that use backbone coordinate-derived features show exceptional performance on native sequence recovery tasks and are promising frameworks for design. A statistical framework for modeling protein sequence landscapes using Tertiary Motifs (TERMs), compact units of recurring structure in proteins, has also demonstrated good performance on protein design tasks. In this work, we investigate the use of TERM-derived data as features in neural protein design frameworks. Our graph-based architecture, TERMinator, incorporates TERM-based and coordinate-based information and outputs a Potts model over sequence space. TERMinator outperforms state-of-the-art models on native sequence recovery tasks, suggesting that utilizing TERM-based and coordinate-based features together is beneficial for protein design.
翻译:计算蛋白质的设计有可能为多种应用提供新型分子结构、粘合器和催化剂。最近以神经图表为基础的模型,使用主干坐标生成的特征,显示了本地序列恢复任务的特殊性,是很有希望的设计框架。一个利用蛋白质中复现结构的紧凑单位 -- -- Terric Motifs(Treets)模拟蛋白质序列景观的统计框架也显示了蛋白质设计任务的良好性能。在这项工作中,我们调查了长期衍生数据作为神经蛋白设计框架特征的使用情况。我们基于图表的结构,即Myterinator,包含了基于Mys的、基于协调的信息和产出。Motts模型超越了空间序列。Motinators在本地序列恢复任务上超越了最先进的模型,表明利用基于My和基于协调的特征一起对蛋白质设计有益。