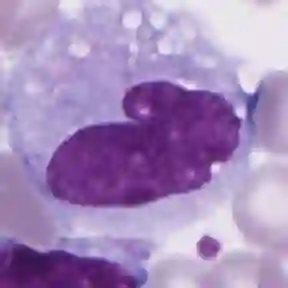
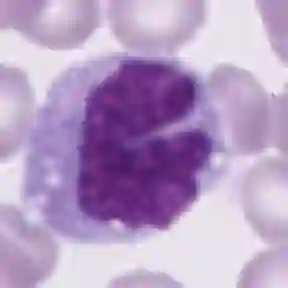
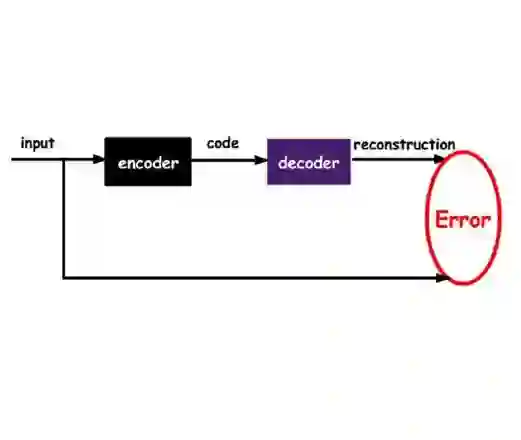
Diagnosing hematological malignancies requires identification and classification of white blood cells in peripheral blood smears. Domain shifts caused by different lab procedures, staining, illumination, and microscope settings hamper the re-usability of recently developed machine learning methods on data collected from different sites. Here, we propose a cross-domain adapted autoencoder to extract features in an unsupervised manner on three different datasets of single white blood cells scanned from peripheral blood smears. The autoencoder is based on an R-CNN architecture allowing it to focus on the relevant white blood cell and eliminate artifacts in the image. To evaluate the quality of the extracted features we use a simple random forest to classify single cells. We show that thanks to the rich features extracted by the autoencoder trained on only one of the datasets, the random forest classifier performs satisfactorily on the unseen datasets, and outperforms published oracle networks in the cross-domain task. Our results suggest the possibility of employing this unsupervised approach in more complicated diagnosis and prognosis tasks without the need to add expensive expert labels to unseen data.
翻译:血清恶性诊断要求确认和分类周边血液涂片中的白血细胞。 不同实验室程序、 污点、 光化和显微镜设置导致的域变妨碍最近开发的关于从不同地点收集的数据的机器学习方法的可重新使用性。 这里, 我们提出一个跨域经改造的自动编码器, 以不受监督的方式提取从周边血液涂片中扫描的单一白细胞的三个不同数据集的特征。 自动编码器基于一个 R- CNN 结构, 使其能够关注相关的白血细胞, 并消除图像中的文物。 为了评估提取的特征的质量, 我们使用简单的随机森林来对单个细胞进行分类。 我们表明,由于只对一个数据集进行自动编码培训的自动编码器所提取的丰富特征, 随机的森林分类器在未知的数据集上表现令人满意, 超越了跨域任务中已公布的孔径网络。 我们的结果表明, 有可能在更复杂的诊断和剖析工作中采用这种未超的处理方法, 而不需要增加昂贵的专家标签。