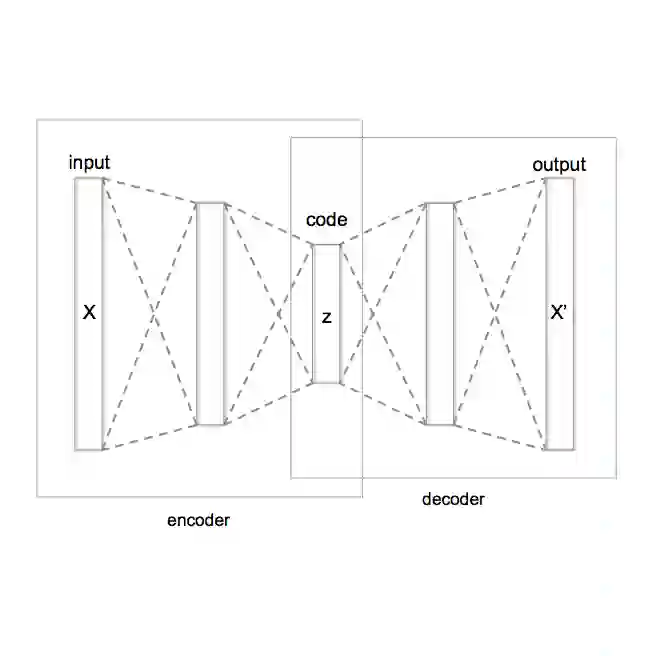
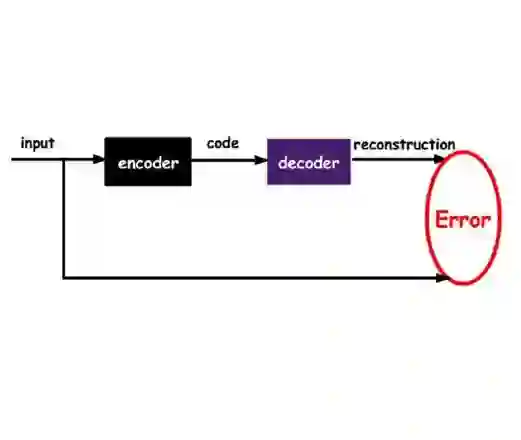
The application of deep learning to generative molecule design has shown early promise for accelerating lead series development. However, questions remain concerning how factors like training, dataset, and seed bias impact the technology's utility to medicine and computational chemists. In this work, we analyze the impact of seed and training bias on the output of an activity-conditioned graph-based variational autoencoder (VAE). Leveraging a massive, labeled dataset corresponding to the dopamine D2 receptor, our graph-based generative model is shown to excel in producing desired conditioned activities and favorable unconditioned physical properties in generated molecules. We implement an activity swapping method that allows for the activation, deactivation, or retention of activity of molecular seeds, and we apply independent deep learning classifiers to verify the generative results. Overall, we uncover relationships between noise, molecular seeds, and training set selection across a range of latent-space sampling procedures, providing important insights for practical AI-driven molecule generation.
翻译:将深层次学习应用于基因分子设计,显示了加速铅序列开发的早期前景;然而,关于培训、数据集和种子偏见等因素如何影响该技术对医药和计算化学家的效用,仍然存在一些问题。在这项工作中,我们分析了种子和培训偏见对活动条件图形化变异自动编码器(VAE)产出的影响。利用与多巴胺D2受体相对的大规模标签数据集,我们的图基基因模型显示在创造所需条件活动和生成分子中有利的无附加条件物理特性方面优异。我们采用了一种活动交换方法,允许激活、停用或保留分子种子的活动,我们运用独立的深层学习分类器来核查基因化结果。总体而言,我们发现在一系列潜空取样程序中噪音、分子种子和培训设置选择之间的关系,为实用的由AI驱动的分子生成提供了重要的洞察力。