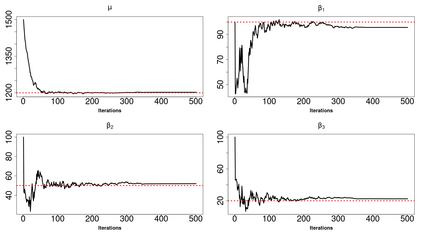
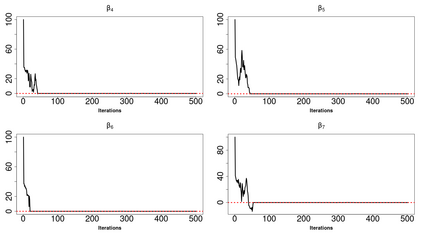
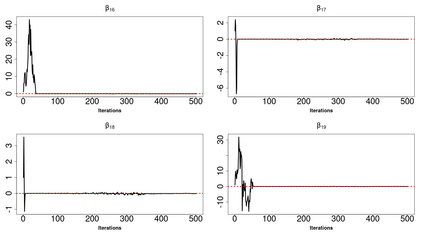
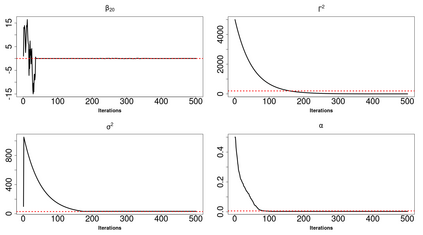
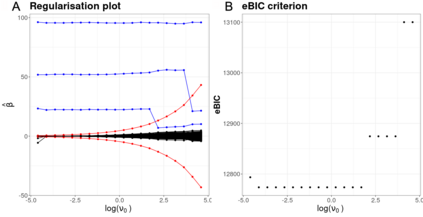
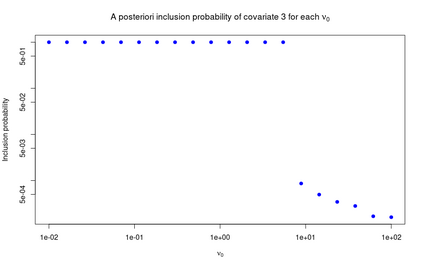
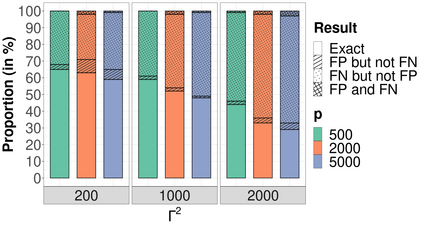
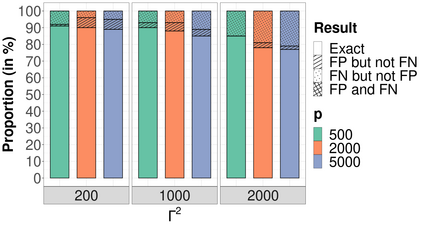
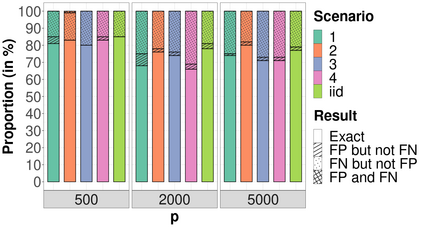
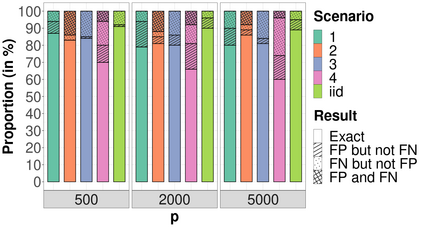
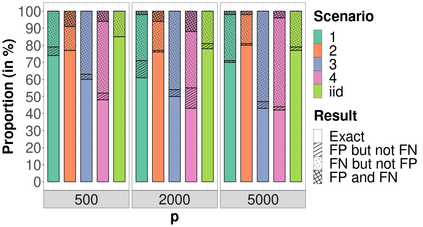
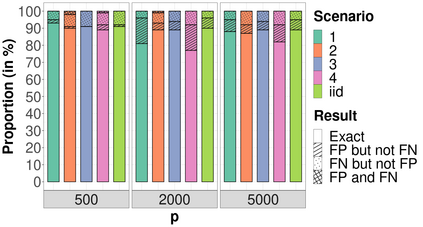
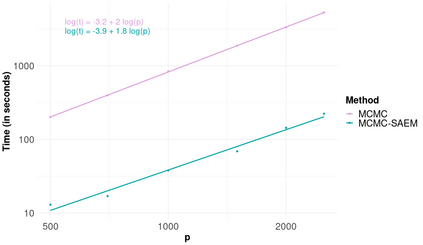
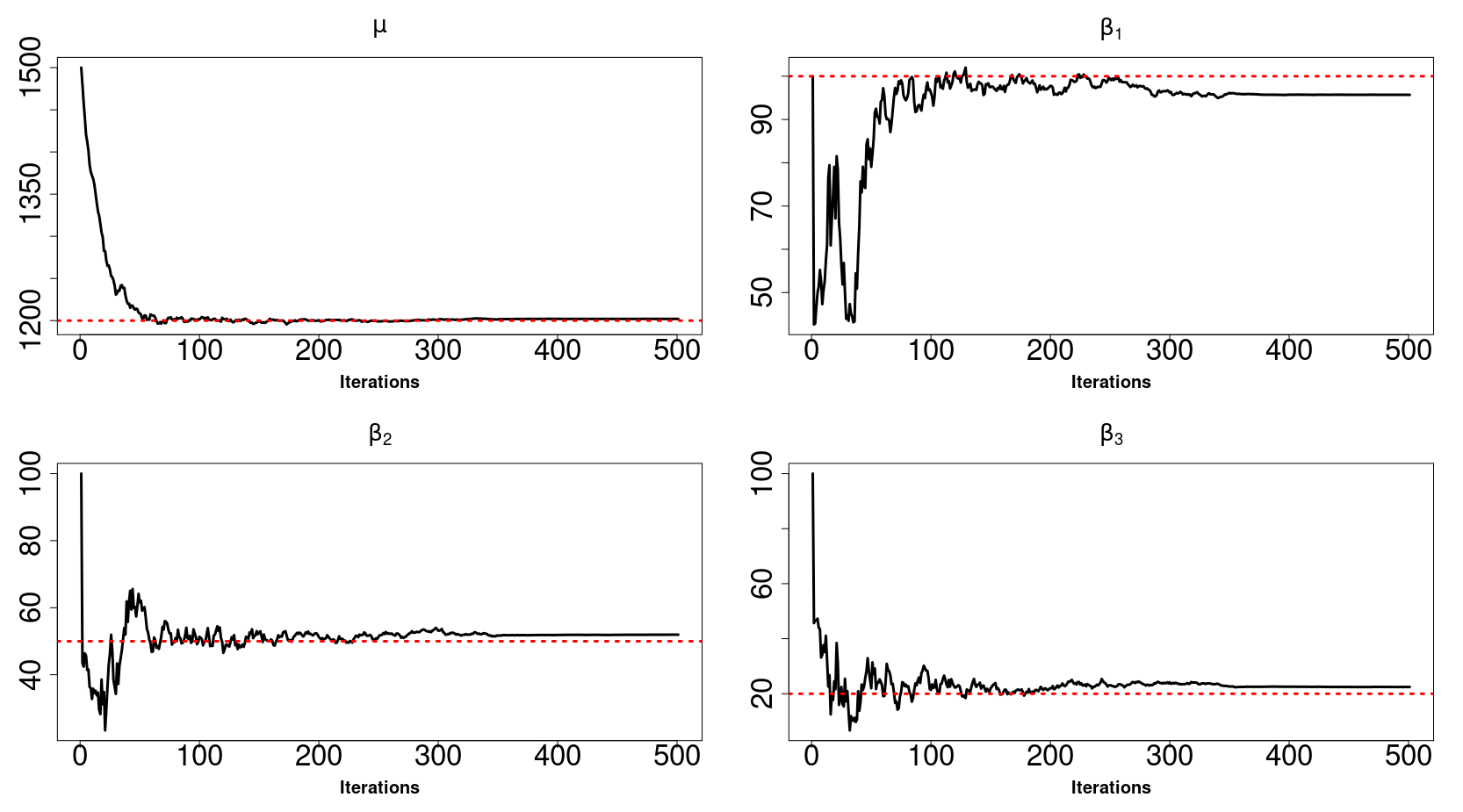
High-dimensional data, with many more covariates than observations, such as genomic data for example, are now commonly analysed. In this context, it is often desirable to be able to focus on the few most relevant covariates through a variable selection procedure. High-dimensional variable selection is widely documented in standard regression models, but there are still few tools to address it in the context of non-linear mixed-effects models. In this work, variable selection is approached from a Bayesian perspective and a selection procedure is proposed, combining the use of spike-and-slab priors and the SAEM algorithm. Similarly to LASSO regression, the set of relevant covariates is selected by exploring a grid of values for the penalisation parameter. The proposed approach is much faster than a classical MCMC algorithm and shows very good selection performances on simulated data.
翻译:高维数据比观测数据(例如基因组数据)多得多的共变数据,现在通常得到分析。在这方面,通常最好能够通过一个可变选择程序集中关注少数最相关的共变数据。高维变量选择在标准回归模型中广泛记录,但在非线性混合效应模型中仍然没有什么工具可以解决这个问题。在这项工作中,从巴伊西亚角度选择变量,并提议了一个选择程序,结合使用钉钉和丝带前置法以及SAEM算法。与LASSO回归法相似,相关的共变数组是通过探索惩罚参数的值网选择的。拟议方法比传统的MCMC算法要快得多,在模拟数据上显示非常良好的选择性能。