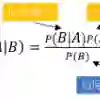
In today's data-intensive landscape, where high-dimensional datasets are increasingly common, reducing the number of input features is essential to prevent overfitting and improve model accuracy. Despite numerous efforts to tackle dimensionality reduction, most approaches apply a universal set of features across all classes, potentially missing the unique characteristics of individual classes. This paper presents the Explainable Class-Specific Naive Bayes (XNB) classifier, which introduces two critical innovations: 1) the use of Kernel Density Estimation to calculate posterior probabilities, allowing for a more accurate and flexible estimation process, and 2) the selection of class-specific feature subsets, ensuring that only the most relevant variables for each class are utilized. Extensive empirical analysis on high-dimensional genomic datasets shows that XNB matches the classification performance of traditional Naive Bayes while drastically improving model interpretability. By isolating the most relevant features for each class, XNB not only reduces the feature set to a minimal, distinct subset for each class but also provides deeper insights into how the model makes predictions. This approach offers significant advantages in fields where both precision and explainability are critical.
翻译:暂无翻译