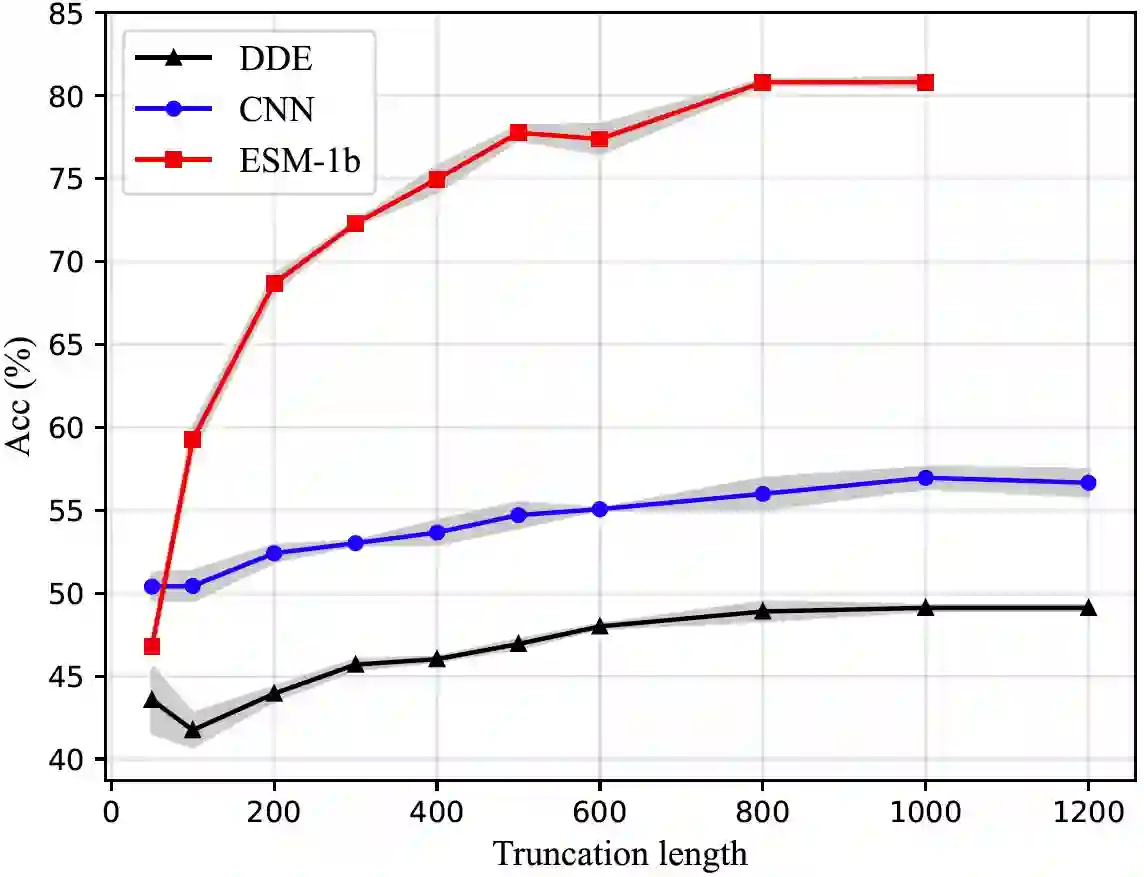
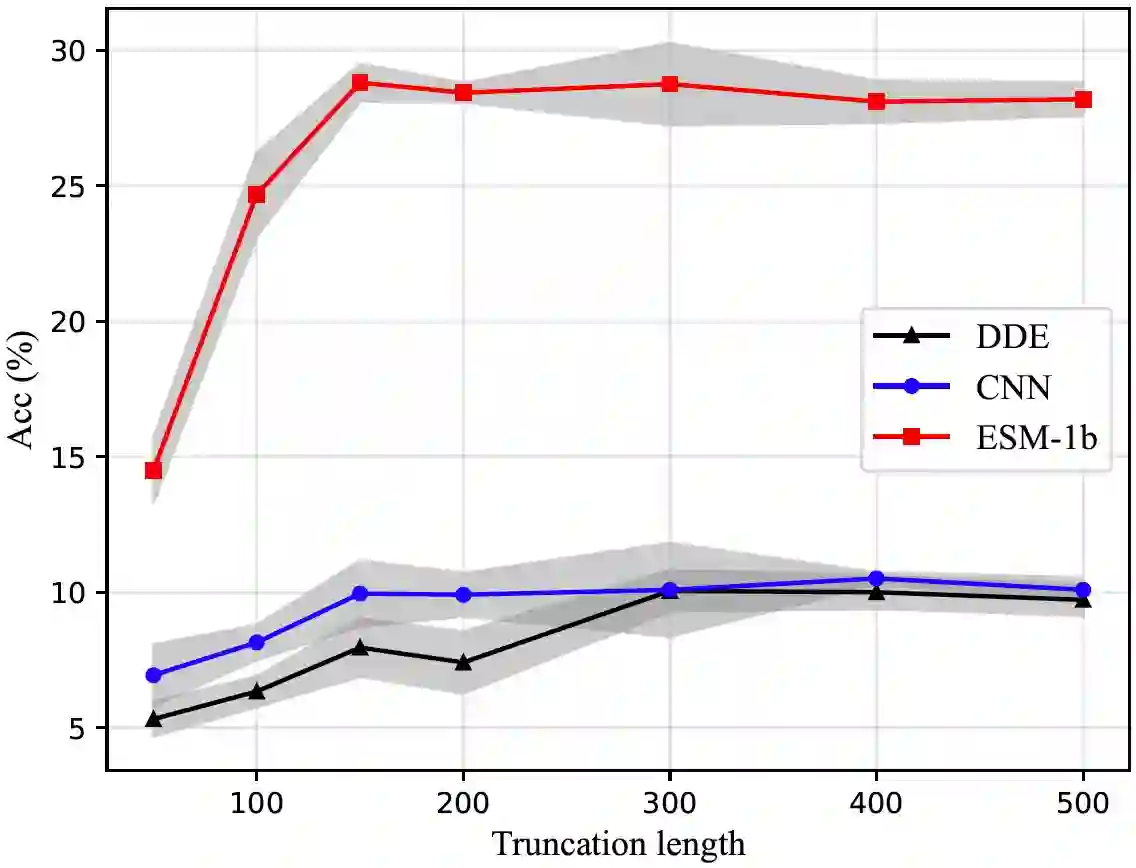
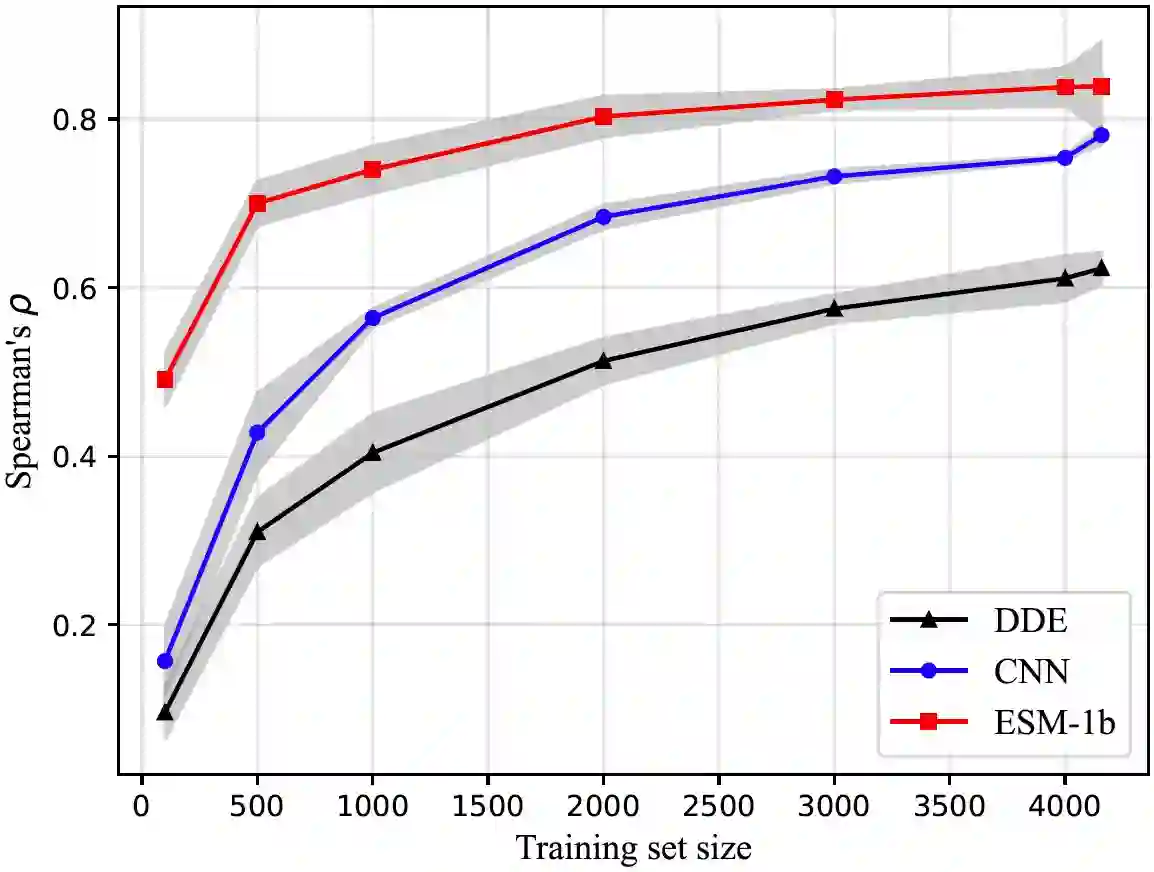
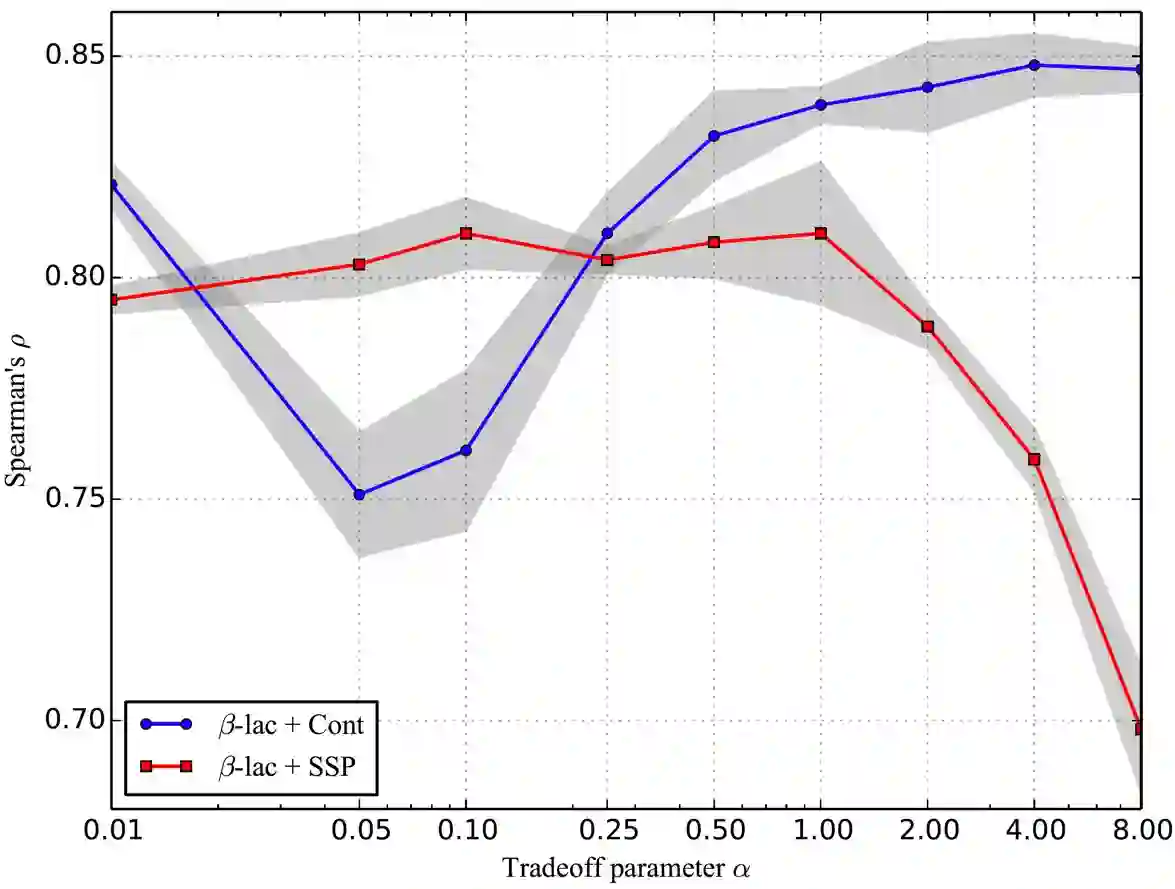
We are now witnessing significant progress of deep learning methods in a variety of tasks (or datasets) of proteins. However, there is a lack of a standard benchmark to evaluate the performance of different methods, which hinders the progress of deep learning in this field. In this paper, we propose such a benchmark called PEER, a comprehensive and multi-task benchmark for Protein sEquence undERstanding. PEER provides a set of diverse protein understanding tasks including protein function prediction, protein localization prediction, protein structure prediction, protein-protein interaction prediction, and protein-ligand interaction prediction. We evaluate different types of sequence-based methods for each task including traditional feature engineering approaches, different sequence encoding methods as well as large-scale pre-trained protein language models. In addition, we also investigate the performance of these methods under the multi-task learning setting. Experimental results show that large-scale pre-trained protein language models achieve the best performance for most individual tasks, and jointly training multiple tasks further boosts the performance. The datasets and source codes of this benchmark are all available at https://github.com/DeepGraphLearning/PEER_Benchmark
翻译:目前,在各种蛋白质的任务(或数据集)中,深层学习方法取得了显著进展,然而,缺乏评价不同方法绩效的标准基准,这阻碍了该领域深层学习的进展。在本文件中,我们提议了一个称为PEER(Protein sEquation understand)的综合和多任务基准,即蛋白质质量综合和多任务基准。PEER提供了一系列不同的蛋白质理解任务,包括蛋白质功能预测、蛋白质本地化预测、蛋白质结构预测、蛋白质互动预测和蛋白质-骨和互动预测。我们评估了每种任务的不同类型基于序列的方法,包括传统特征工程方法、不同序列编码方法以及大规模预先培训的蛋白语言模型。此外,我们还调查了这些方法在多任务学习环境中的绩效。实验结果表明,大规模预先培训的蛋白语言模型在大多数单个任务中都取得了最佳性能,联合培训多项任务,进一步提高了绩效。这一基准的数据集和源代码都可在 https://githhubub.com/DeepLasion/sionalveinstagesion。