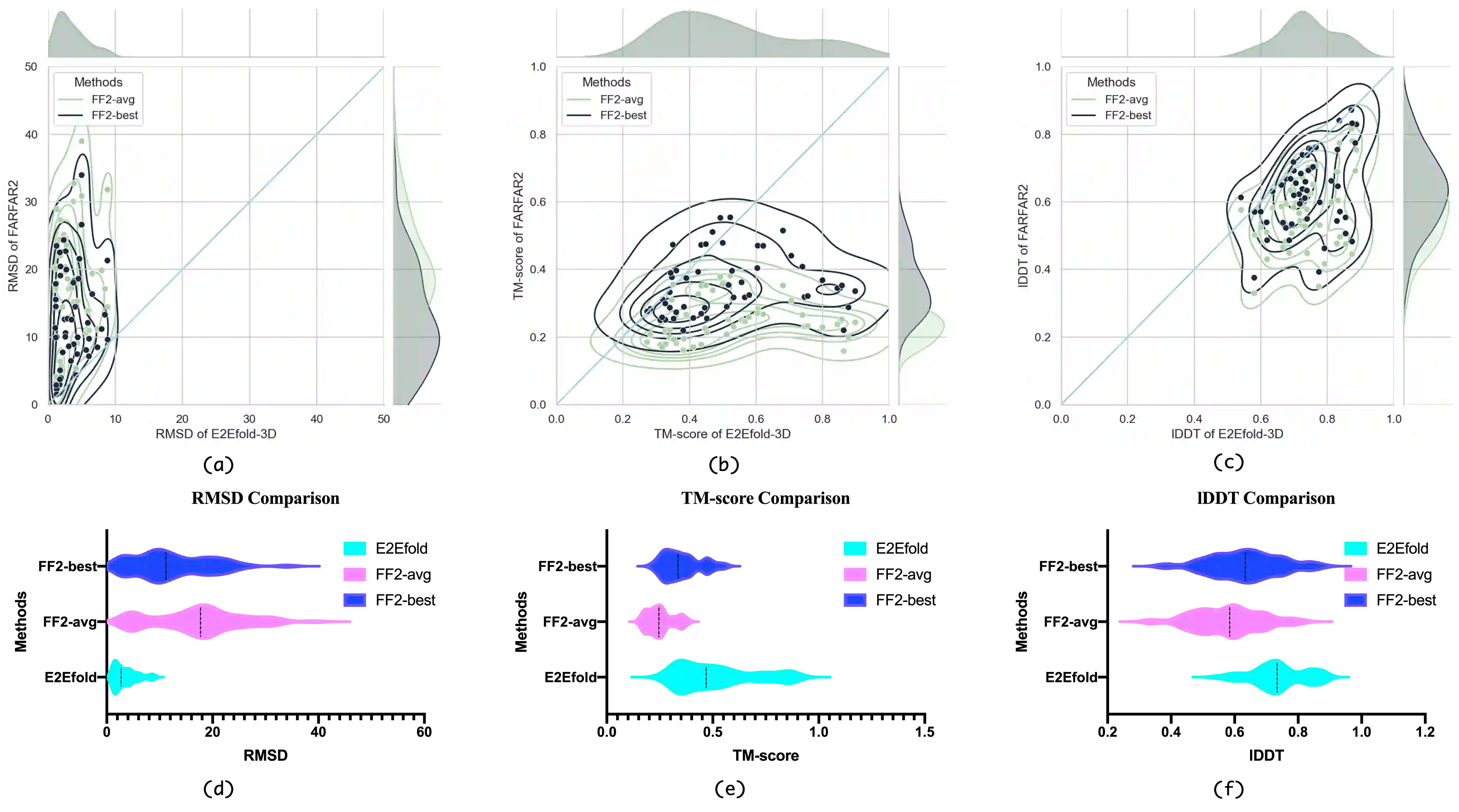
RNA structure determination and prediction can promote RNA-targeted drug development and engineerable synthetic elements design. But due to the intrinsic structural flexibility of RNAs, all the three mainstream structure determination methods (X-ray crystallography, NMR, and Cryo-EM) encounter challenges when resolving the RNA structures, which leads to the scarcity of the resolved RNA structures. Computational prediction approaches emerge as complementary to the experimental techniques. However, none of the \textit{de novo} approaches is based on deep learning since too few structures are available. Instead, most of them apply the time-consuming sampling-based strategies, and their performance seems to hit the plateau. In this work, we develop the first end-to-end deep learning approach, E2Efold-3D, to accurately perform the \textit{de novo} RNA structure prediction. Several novel components are proposed to overcome the data scarcity, such as a fully-differentiable end-to-end pipeline, secondary structure-assisted self-distillation, and parameter-efficient backbone formulation. Such designs are validated on the independent, non-overlapping RNA puzzle testing dataset and reach an average sub-4 \AA{} root-mean-square deviation, demonstrating its superior performance compared to state-of-the-art approaches. Interestingly, it also achieves promising results when predicting RNA complex structures, a feat that none of the previous systems could accomplish. When E2Efold-3D is coupled with the experimental techniques, the RNA structure prediction field can be greatly advanced.
翻译:RNA结构的确定和预测可以促进RNA针对药物的开发和可工程师合成元素的设计。但是,由于RNA的内在结构灵活性,所有三个主流结构的确定方法(X-光晶晶体学、NMR和Cryo-EM)在解决RNA结构时都遇到挑战,这导致解决RNA结构的缺乏。计算预测方法作为实验技术的补充出现。然而,没有一个计算方法是基于深层次学习的,因为结构太少。相反,它们大多采用耗时抽样战略,其性能似乎达到顶峰值。在这项工作中,我们开发了第一个端到端的深层次学习方法,即E2Efoldy-3D,以准确地进行已解决的RNA结构结构。提出了若干新的组成部分,以克服数据短缺,例如完全不同的端到端的管道,二级结构辅助的自我蒸馏,以及参数高效的主干配方。这种设计在独立、非重叠的抽样抽样战略上验证了RNA-3技术,其最终结果也表明其深度的精确度,其深度的精确性根基值的精确性数据测试结果。