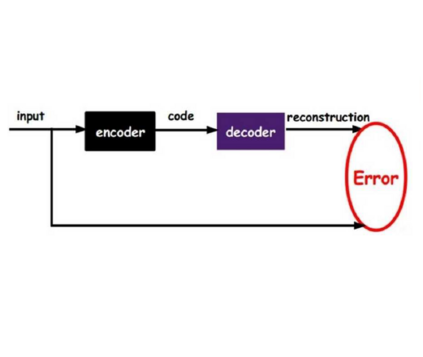
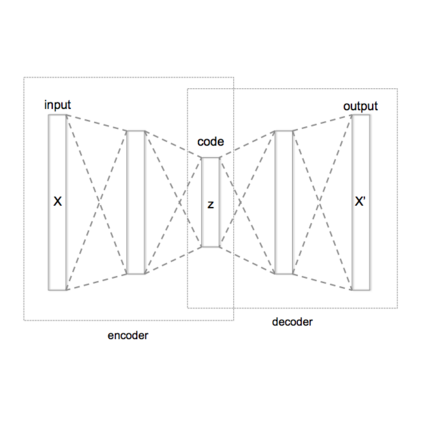
Probabilistic generative models are attractive for scientific modeling because their inferred parameters can be used to generate hypotheses and design experiments. This requires that the learned model provide an accurate representation of the input data and yield a latent space that effectively predicts outcomes relevant to the scientific question. Supervised Variational Autoencoders (SVAEs) have previously been used for this purpose, where a carefully designed decoder can be used as an interpretable generative model while the supervised objective ensures a predictive latent representation. Unfortunately, the supervised objective forces the encoder to learn a biased approximation to the generative posterior distribution, which renders the generative parameters unreliable when used in scientific models. This issue has remained undetected as reconstruction losses commonly used to evaluate model performance do not detect bias in the encoder. We address this previously-unreported issue by developing a second order supervision framework (SOS-VAE) that influences the decoder to induce a predictive latent representation. This ensures that the associated encoder maintains a reliable generative interpretation. We extend this technique to allow the user to trade-off some bias in the generative parameters for improved predictive performance, acting as an intermediate option between SVAEs and our new SOS-VAE. We also use this methodology to address missing data issues that often arise when combining recordings from multiple scientific experiments. We demonstrate the effectiveness of these developments using synthetic data and electrophysiological recordings with an emphasis on how our learned representations can be used to design scientific experiments.
翻译:概率感测模型对科学模型具有吸引力,因为其推论参数可用于产生假设和设计实验。这要求所学模型能够准确反映输入数据,并产生一个潜在空间,从而有效地预测与科学问题有关的结果。以前曾为此目的使用过监督的变异自动编码器(SVAE),在此过程中,可以使用精心设计的解调器作为可解释的基因模型,而监督的目标则确保了预测的潜在代表性。不幸的是,受监督的目标强调迫使编码器学习对基因化远地点分布的偏差近,从而使基因化参数在科学模型中使用时变得不可靠。由于通常用来评价模型性能的重建损失没有察觉到编码器中的偏差,这一问题一直没有被察觉到。我们以前未报告过的问题是通过开发第二个可解释的调控框架(SOS-VAE)来解决这个问题,它影响解析器以产生预测性潜在代表性。这确保相关的解析器保持可靠的遗传学解释。我们把这一技术从让用户在使用基因模型时,将某种变异性参数推广到使用一种中间性数据模拟性设计方法,同时使用这种数据模拟模拟模拟模拟的模拟数据模拟模拟模拟数据模拟模拟模拟模拟模拟数据,用以显示方法。我们用这种方法处理这种数据的模拟模拟模拟数据模拟模拟模拟的模拟模拟模拟的模拟数据模拟数据模拟数据模拟数据模拟数据模拟的演化方法处理。