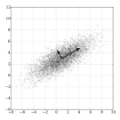
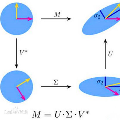
Principal components computed via PCA (principal component analysis) are traditionally used to reduce dimensionality in genomic data or to correct for population stratification. In this paper, we explore the penalized eigenvalue problem (PEP) which reformulates the computation of the first eigenvector as an optimization problem and adds an L1 penalty constraint. The contribution of our article is threefold. First, we extend PEP by applying Nesterov smoothing to the original LASSO-type L1 penalty. This allows one to compute analytical gradients which enable faster and more efficient minimization of the objective function associated with the optimization problem. Second, we demonstrate how higher order eigenvectors can be calculated with PEP using established results from singular value decomposition (SVD). Third, using data from the 1000 Genome Project dataset, we empirically demonstrate that our proposed smoothed PEP allows one to increase numerical stability and obtain meaningful eigenvectors. We further investigate the utility of the penalized eigenvector approach over traditional PCA.
翻译:暂无翻译