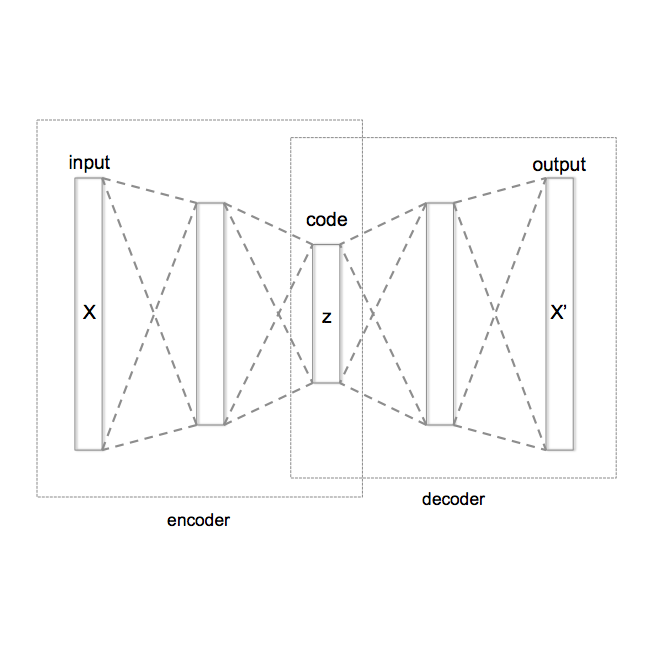
Much scientific enquiry across disciplines is founded upon a mechanistic treatment of dynamic systems that ties form to function. A highly visible instance of this is in molecular biology, where an important goal is to determine functionally-relevant forms/structures that a protein molecule employs to interact with molecular partners in the living cell. This goal is typically pursued under the umbrella of stochastic optimization with algorithms that optimize a scoring function. Research repeatedly shows that current scoring function, though steadily improving, correlate weakly with molecular activity. Inspired by recent momentum in generative deep learning, this paper proposes and evaluates an alternative approach to generating functionally-relevant three-dimensional structures of a protein. Though typically deep generative models struggle with highly-structured data, the work presented here circumvents this challenge via graph-generative models. A comprehensive evaluation of several deep architectures shows the promise of generative models in directly revealing the latent space for sampling novel tertiary structures, as well as in highlighting axes/factors that carry structural meaning and open the black box often associated with deep models. The work presented here is a first step towards interpretative, deep generative models becoming viable and informative complementary approaches to protein structure prediction.
翻译:大量学科间科学调查基于对与功能相关联的动态系统的机械处理。在分子生物学中,这是一个非常明显的事例,其重要目标是确定一个蛋白质分子与活细胞中的分子伙伴互动的功能相关形式/结构。这个目标通常是在随机优化和优化评分功能的算法的组合下追求的。研究一再表明,目前的评分功能虽然稳步改善,但与分子活动关系不大。在基因深层次学习的最新势头的启发下,本文件提出并评估了产生与功能相关的蛋白质三维结构的替代方法。尽管典型的深层基因化模型与高度结构化的数据挣扎,但这里介绍的工作通过图形生成模型回避了这一挑战。对一些深层结构的全面评价表明,在直接揭示取样新大学结构的潜在空间方面,以及在突出具有结构性意义和打开往往与深层模型相关的黑盒的轴/因素方面,基因深层次的模型是走向解释性、深层次基因化模型成为对蛋白质结构的可行和丰富性补充性预测的第一步。