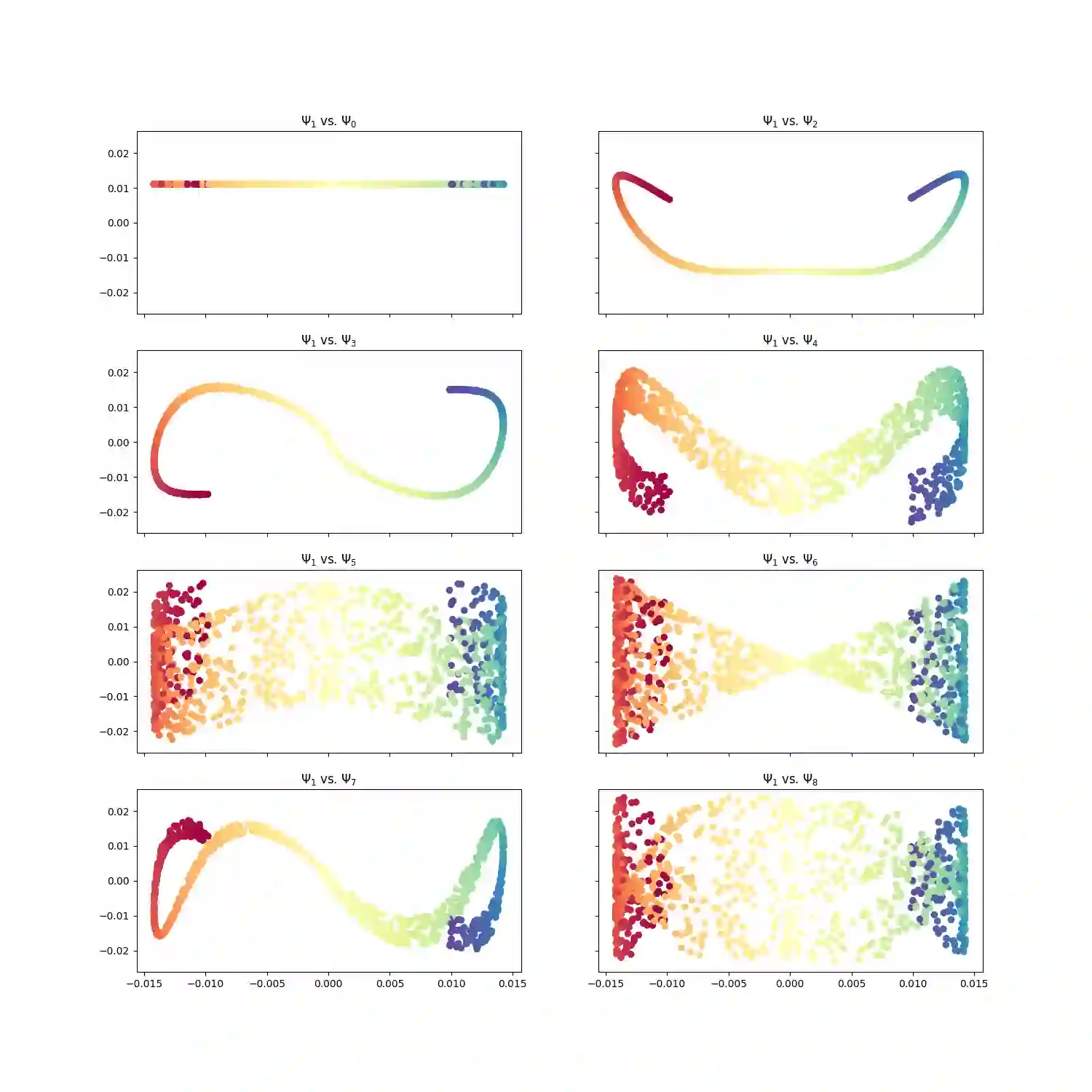
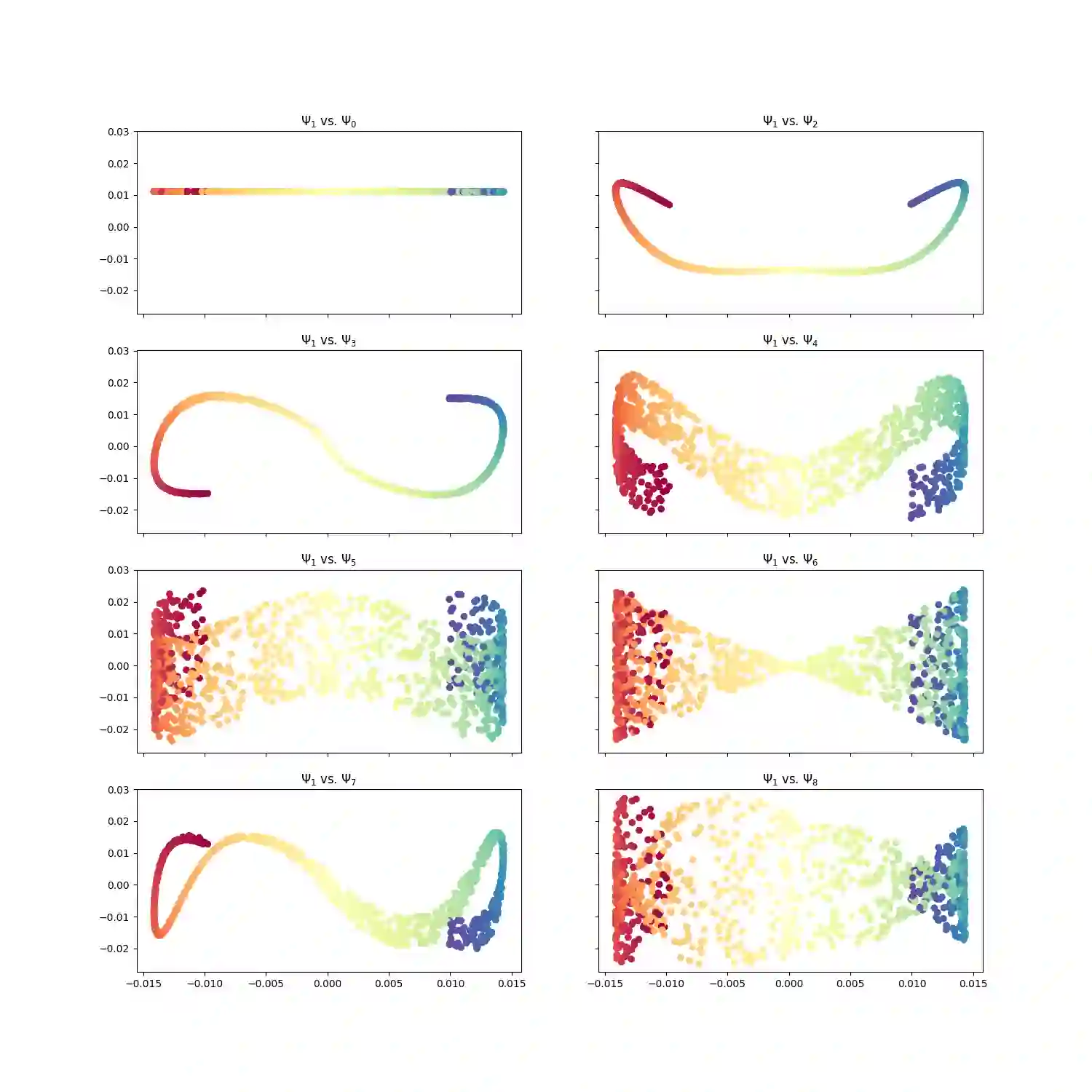
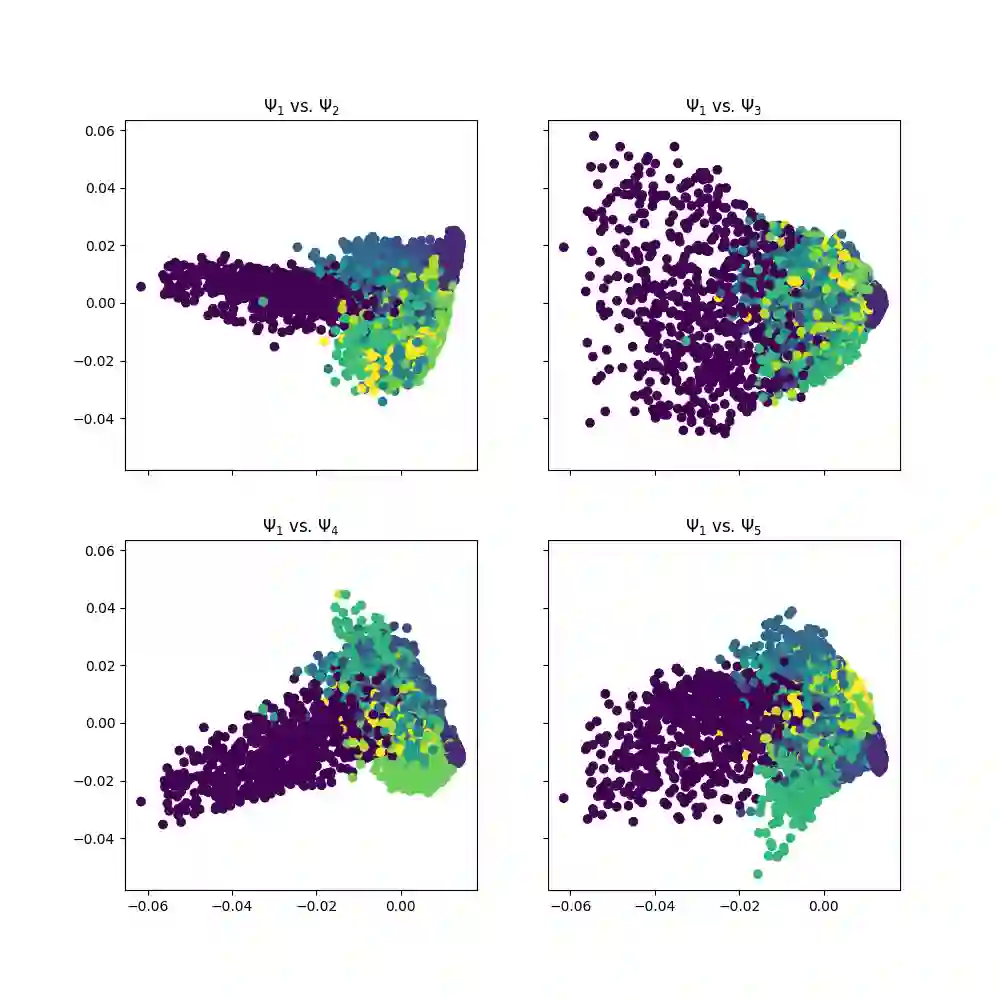
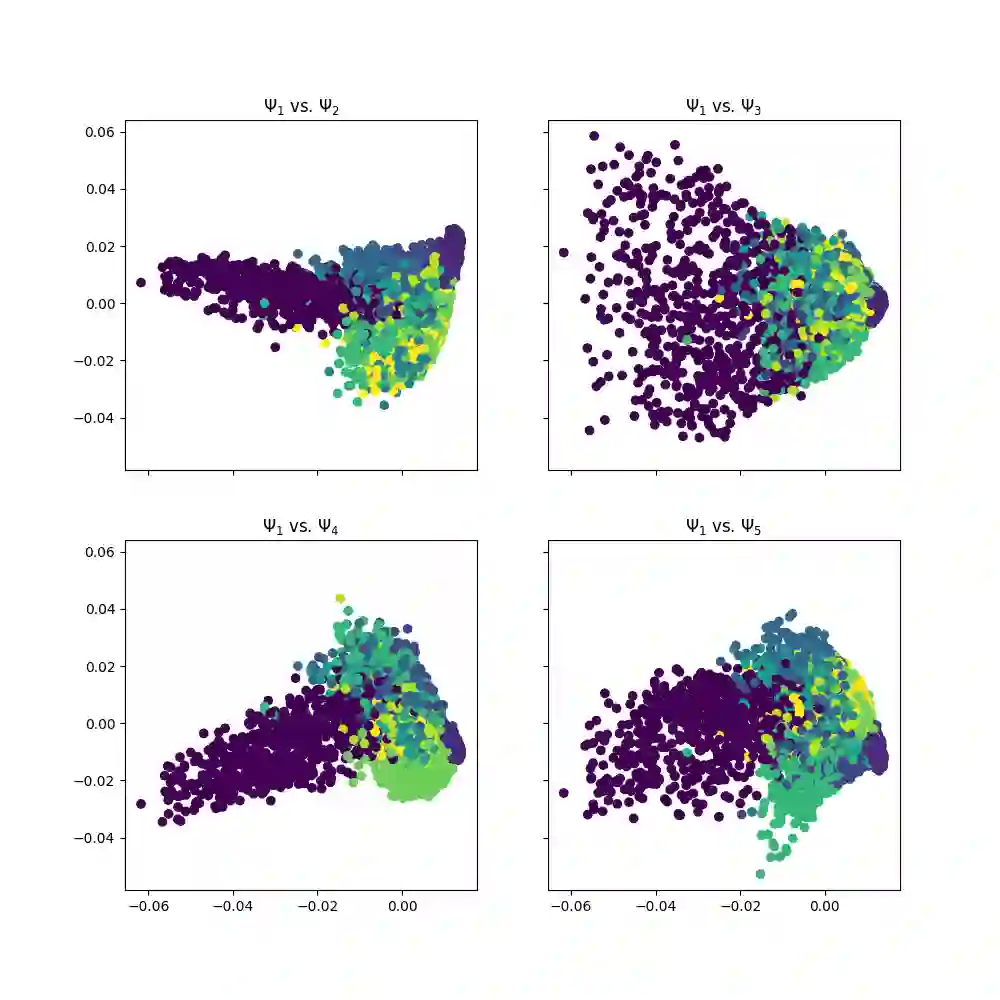
With the emergence of Artificial Intelligence, numerical algorithms are moving towards more approximate approaches. For methods such as PCA or diffusion maps, it is necessary to compute eigenvalues of a large matrix, which may also be dense depending on the kernel. A global method, i.e. a method that requires all data points simultaneously, scales with the data dimension N and not with the intrinsic dimension d; the complexity for an exact dense eigendecomposition leads to $\mathcal{O}(N^{3})$. We have combined the two frameworks, $\mathsf{datafold}$ and $\mathsf{GOFMM}$. The first framework computes diffusion maps, where the computational bottleneck is the eigendecomposition while with the second framework we compute the eigendecomposition approximately within the iterative Lanczos method. A hierarchical approximation approach scales roughly with a runtime complexity of $\mathcal{O}(Nlog(N))$ vs. $\mathcal{O}(N^{3})$ for a classic approach. We evaluate the approach on two benchmark datasets -- scurve and MNIST -- with strong and weak scaling using OpenMP and MPI on dense matrices with maximum size of $100k\times100k$.
翻译:暂无翻译