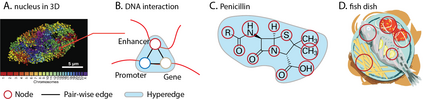
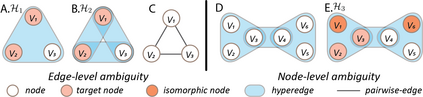
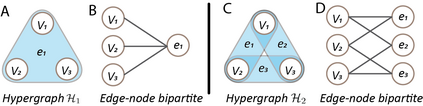
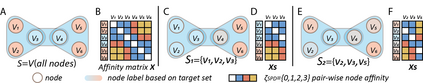
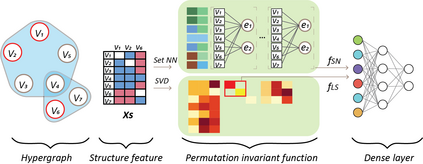
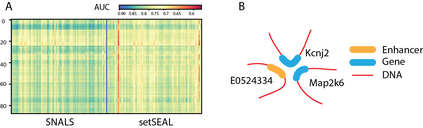
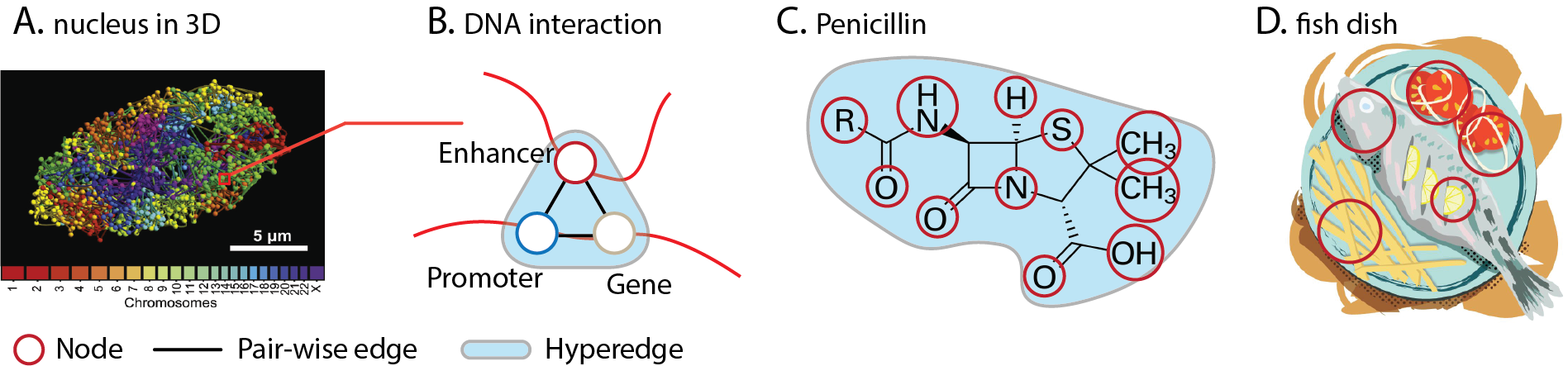
Hypergraph offers a framework to depict the multilateral relationships in real-world complex data. Predicting higher-order relationships, i.e hyperedge, becomes a fundamental problem for the full understanding of complicated interactions. The development of graph neural network (GNN) has greatly advanced the analysis of ordinary graphs with pair-wise relations. However, these methods could not be easily extended to the case of hypergraph. In this paper, we generalize the challenges of GNN in representing higher-order data in principle, which are edge- and node-level ambiguities. To overcome the challenges, we present SNALS that utilizes bipartite graph neural network with structural features to collectively tackle the two ambiguity issues. SNALS captures the joint interactions of a hyperedge by its local environment, which is retrieved by collecting the spectrum information of their connections. As a result, SNALS achieves nearly 30% performance increase compared with most recent GNN-based models. In addition, we applied SNALS to predict genetic higher-order interactions on 3D genome organization data. SNALS showed consistently high prediction accuracy across different chromosomes, and generated novel findings on 4-way gene interaction, which is further validated by existing literature.
翻译:测深仪为描述现实世界复杂数据的多边关系提供了一个框架。 预测高阶关系, 即高端关系, 成为充分了解复杂相互作用的一个根本问题。 图形神经网络( GNNN) 的开发极大地推动了对普通图表和双向关系的分析。 但是,这些方法无法轻易推广到高光谱中。 在本文中, 我们概括了GNN在原则上代表更高阶数据方面的挑战, 它们是边缘和节点层次的模糊性。 为了克服挑战, 我们介绍了SNALS, 利用具有结构特征的双面图形神经网络来集体解决两个模糊性问题。 SNALS 捕捉到由本地环境产生的高端联合相互作用, 后者通过收集其连接的频谱信息检索到。 结果, SNALS 与最近的GNN模型相比, 性能增加了近30%。 此外, 我们应用SNALS来预测3D基因组组织的基因组数据上的更高级互动性。 SNALS显示不同染色体之间的连续高预测准确性, 并生成了四路基因互动的新发现, 由现有文献进一步验证。