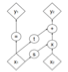
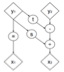
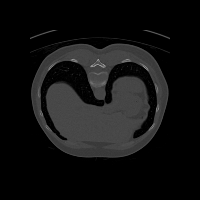
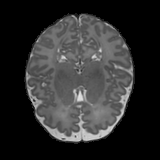
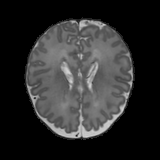
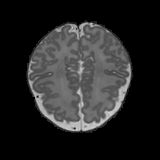
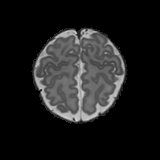
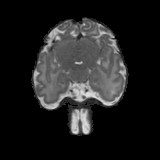
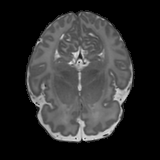
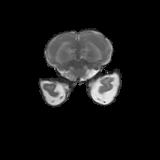
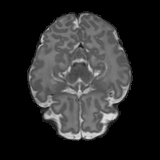
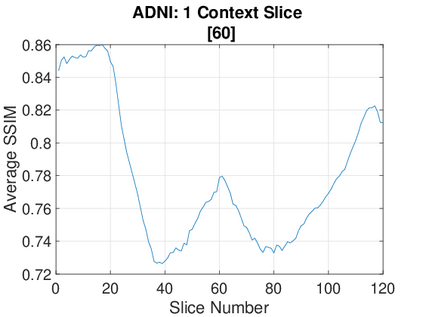
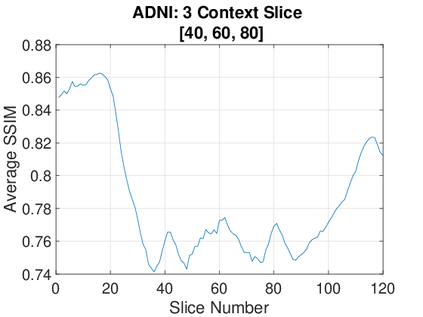
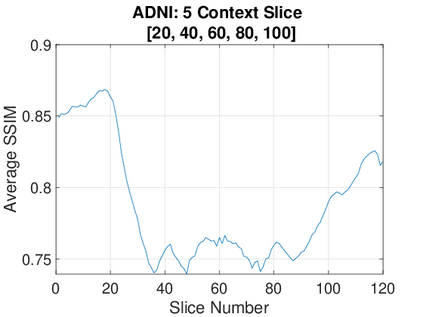
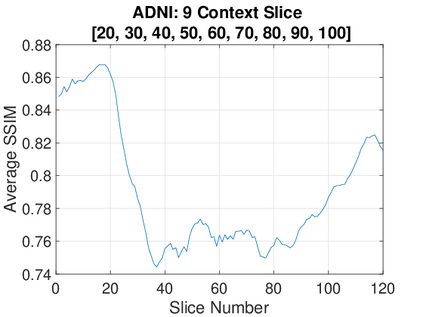
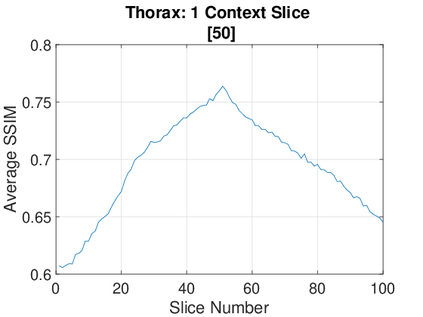
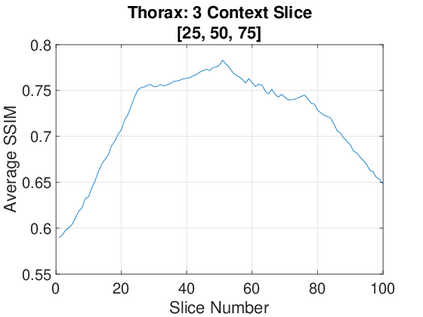
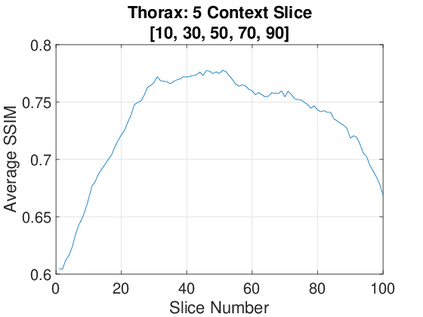
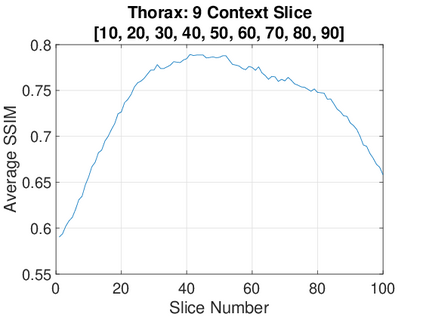
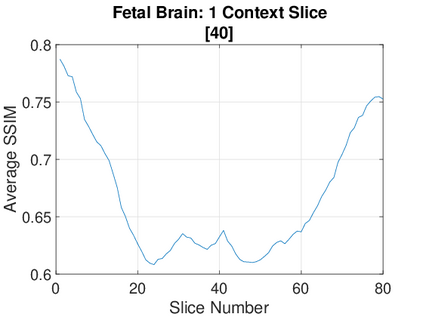
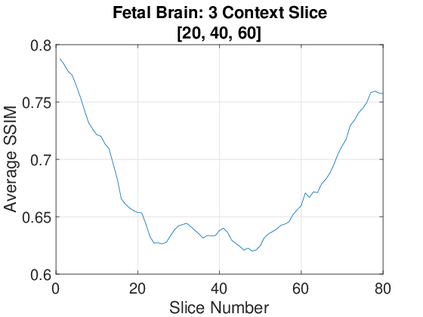
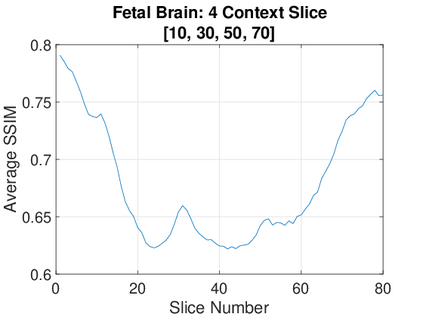
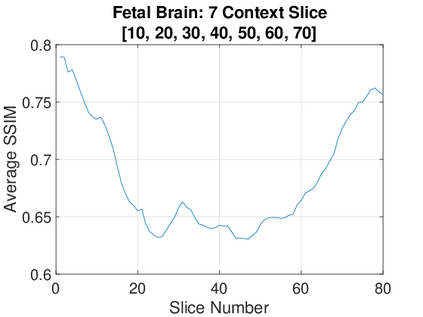
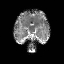
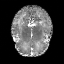
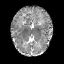
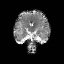
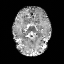
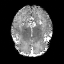
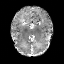
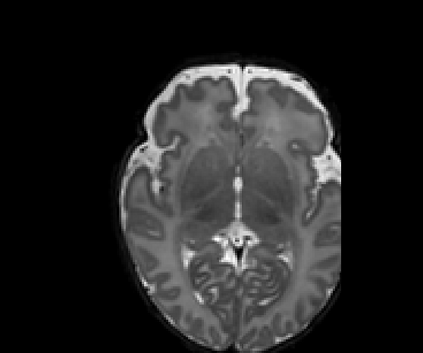
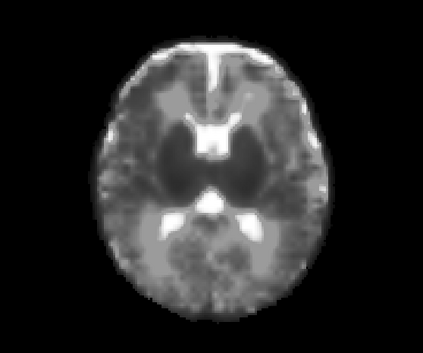
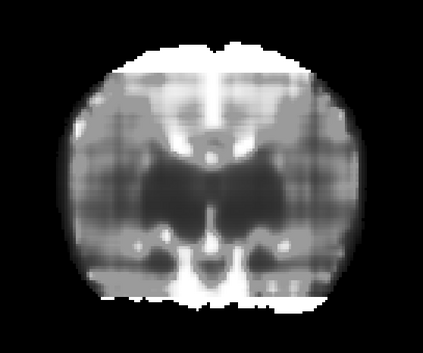
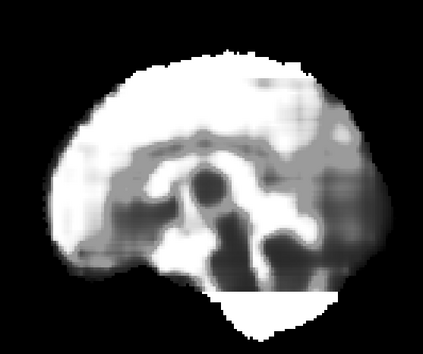
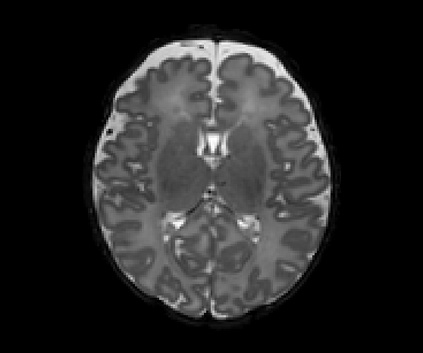
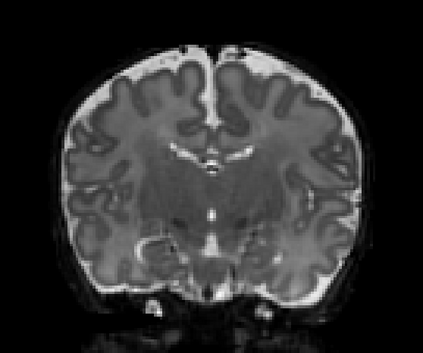
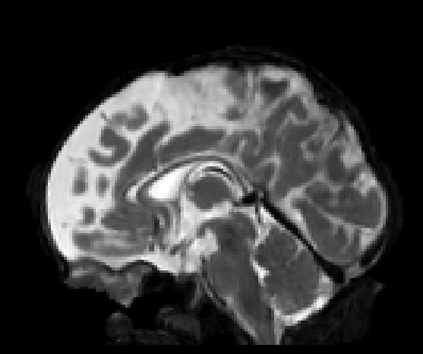
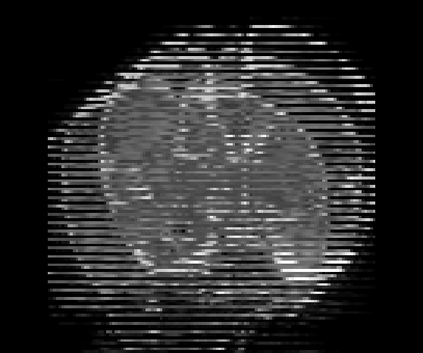
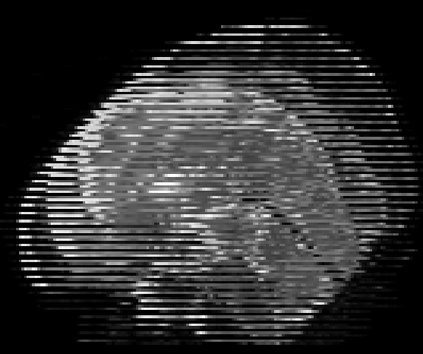
Real-world settings often do not allow acquisition of high-resolution volumetric images for accurate morphological assessment and diagnostic. In clinical practice it is frequently common to acquire only sparse data (e.g. individual slices) for initial diagnostic decision making. Thereby, physicians rely on their prior knowledge (or mental maps) of the human anatomy to extrapolate the underlying 3D information. Accurate mental maps require years of anatomy training, which in the first instance relies on normative learning, i.e. excluding pathology. In this paper, we leverage Bayesian Deep Learning and environment mapping to generate full volumetric anatomy representations from none to a small, sparse set of slices. We evaluate proof of concept implementations based on Generative Query Networks (GQN) and Conditional BRUNO using abdominal CT and brain MRI as well as in a clinical application involving sparse, motion-corrupted MR acquisition for fetal imaging. Our approach allows to reconstruct 3D volumes from 1 to 4 tomographic slices, with a SSIM of 0.7+ and cross-correlation of 0.8+ compared to the 3D ground truth.
翻译:现实世界环境往往不允许获取高分辨率的体积成像,以进行准确的形态评估和诊断;在临床实践中,通常常见的做法是只获得稀少的数据(如个别切片),以进行初步诊断决策;因此,医生依靠其人类解剖学的先前知识(或精神图)来推断基本的3D信息;准确的精神图需要多年的解剖培训,而这种培训首先依靠的是规范学习,即排除病理学;在本文中,我们利用贝耶斯深深层和环境绘图,从零到小、稀少的切片,产生完整的体积解剖面表(如个别切片);我们评估概念实施的证据,其基础是基因质断网(GQN)和致感性BRUNO,使用腹膜CT和大脑MRI,以及临床应用,包括稀疏、运动-腐蚀MRM,用于胎儿成像。我们的方法允许将3D卷从1到4个成像片,用0.7+的SISIM和0.8+的地面与3C的交叉联系。