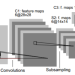
Deep learning has great potential for accurate detection and classification of diseases with medical imaging data, but the performance is often limited by the number of training datasets and memory requirements. In addition, many deep learning models are considered a "black-box," thereby often limiting their adoption in clinical applications. To address this, we present a successive subspace learning model, termed VoxelHop, for accurate classification of Amyotrophic Lateral Sclerosis (ALS) using T2-weighted structural MRI data. Compared with popular convolutional neural network (CNN) architectures, VoxelHop has modular and transparent structures with fewer parameters without any backpropagation, so it is well-suited to small dataset size and 3D imaging data. Our VoxelHop has four key components, including (1) sequential expansion of near-to-far neighborhood for multi-channel 3D data; (2) subspace approximation for unsupervised dimension reduction; (3) label-assisted regression for supervised dimension reduction; and (4) concatenation of features and classification between controls and patients. Our experimental results demonstrate that our framework using a total of 20 controls and 26 patients achieves an accuracy of 93.48$\%$ and an AUC score of 0.9394 in differentiating patients from controls, even with a relatively small number of datasets, showing its robustness and effectiveness. Our thorough evaluations also show its validity and superiority to the state-of-the-art 3D CNN classification methods. Our framework can easily be generalized to other classification tasks using different imaging modalities.
翻译:深层学习具有用医学成像数据对疾病进行准确检测和分类的巨大潜力,但业绩往往受到培训数据集和记忆要求数量的限制。此外,许多深层学习模型被视为“黑盒子”,从而往往限制临床应用中的采用。为此,我们提出一个连续的子空间学习模型,称为VoxelHop,用于对Amyocrictic Late Scleris(ALS)进行准确分类,使用T2加权结构MRI数据。与流行的同源神经网络结构相比,VoxelHop的模块化和透明的结构,其参数较少,没有任何反向分析,因此它完全适合小数据集尺寸和3D成像数据。我们VoxelHop有四个关键组成部分,包括:(1) 连续扩大近距离邻的多频道3D数据;(2) 低超强尺寸的子空间缩缩缩缩缩缩缩;(3) 监督性尺寸降级的标签回归;(4) 控制与病人之间的特征和分类。我们的测试结果显示,我们框架的准确性是完全的194级标准,并用完全的A级和26个病人。