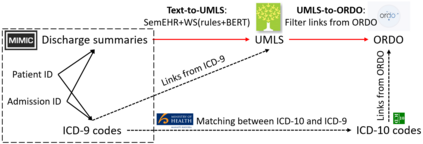
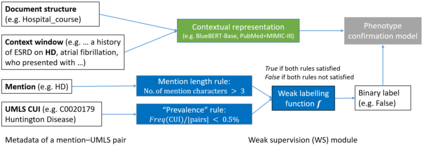
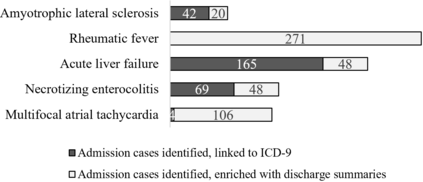
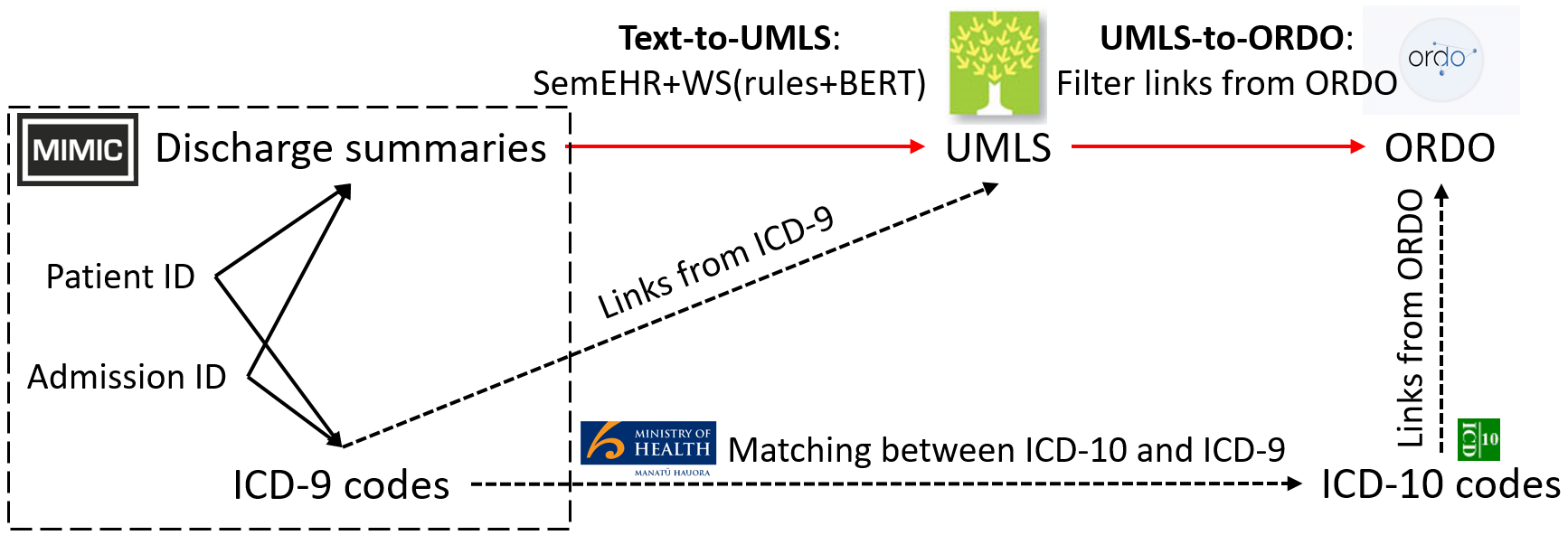
The identification of rare diseases from clinical notes with Natural Language Processing (NLP) is challenging due to the few cases available for machine learning and the need of data annotation from clinical experts. We propose a method using ontologies and weak supervision. The approach includes two steps: (i) Text-to-UMLS, linking text mentions to concepts in Unified Medical Language System (UMLS), with a named entity linking tool (e.g. SemEHR) and weak supervision based on customised rules and Bidirectional Encoder Representations from Transformers (BERT) based contextual representations, and (ii) UMLS-to-ORDO, matching UMLS concepts to rare diseases in Orphanet Rare Disease Ontology (ORDO). Using MIMIC-III US intensive care discharge summaries as a case study, we show that the Text-to-UMLS process can be greatly improved with weak supervision, without any annotated data from domain experts. Our analysis shows that the overall pipeline processing discharge summaries can surface rare disease cases, which are mostly uncaptured in manual ICD codes of the hospital admissions.
翻译:与自然语言处理(NLP)的临床笔记中的稀有病的鉴定具有挑战性,因为机器学习的病例很少,而且需要临床专家提供数据说明。我们提出一种使用本体学和薄弱监督的方法。这个方法包括两个步骤:(一) 文本至UMLS,将提到统一医疗语言系统概念的文字链接到统一医疗语言系统(UMLS)中,有一个名称的实体连接工具(例如SemEHR),根据定制规则进行监管的薄弱,以及基于变异器背景介绍的双向编码显示,以及(二) UMLS至ORDO,将UMLS概念匹配到孤儿病类疾病肿瘤学(ORDO)中的稀有疾病。使用MIMICIII 美国强化护理排放摘要作为案例研究,我们表明文本至UMLS进程可以随着薄弱的监管而大大改进,而没有来自域专家的任何附加说明的数据。我们的分析表明,总管道处理排放摘要可以显示稀有病例病例,而这些病例大多未在医院入手动的ICD编码中。