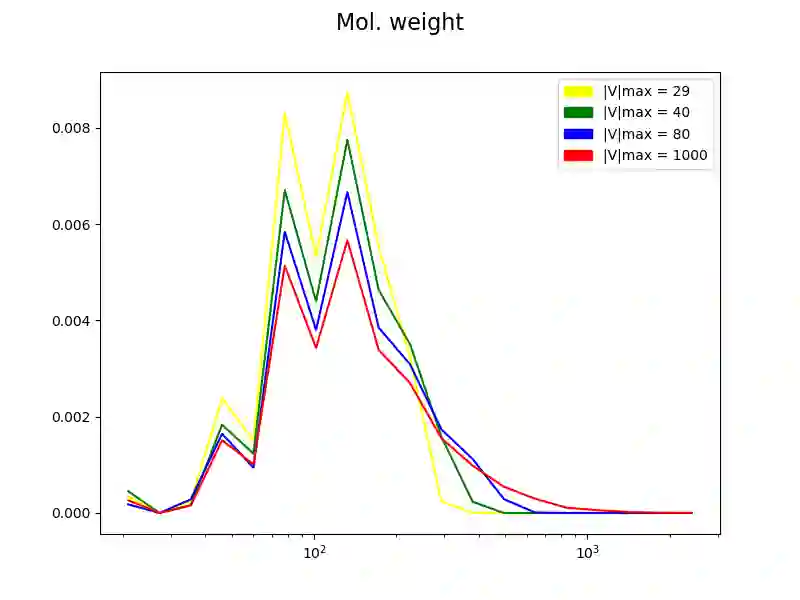
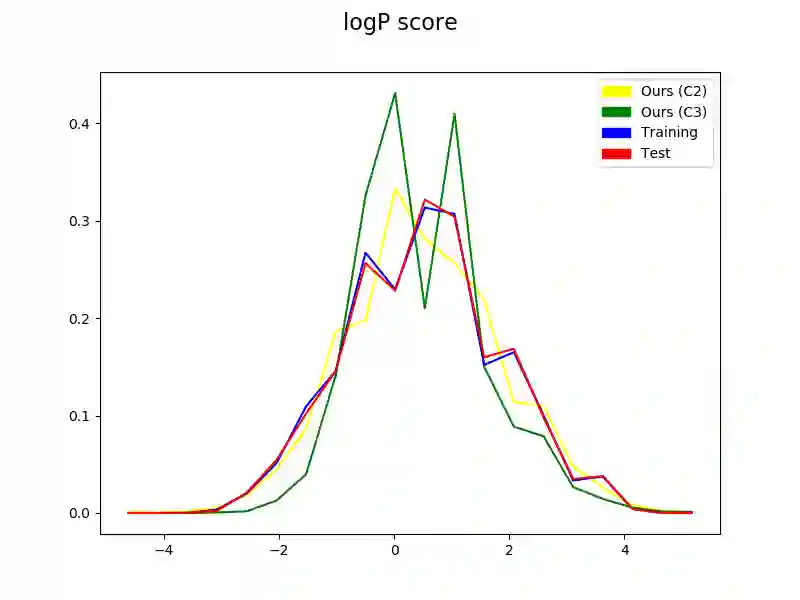
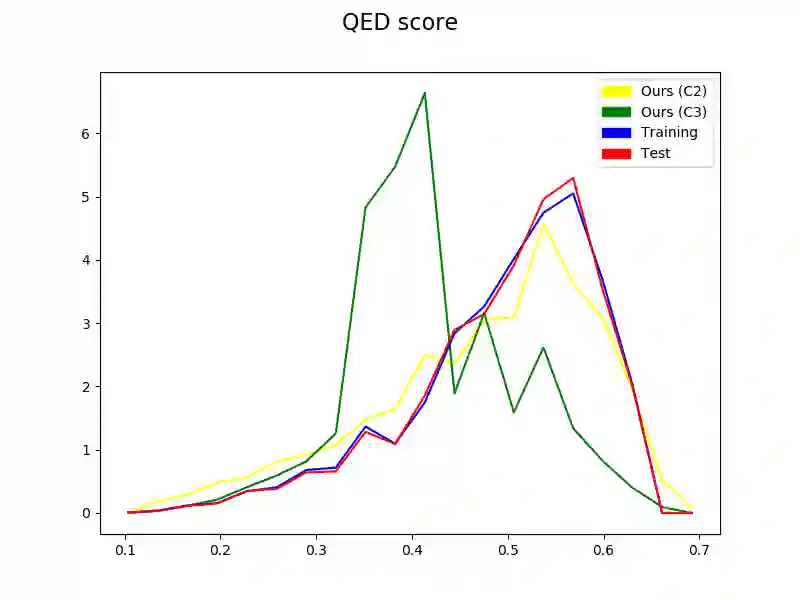
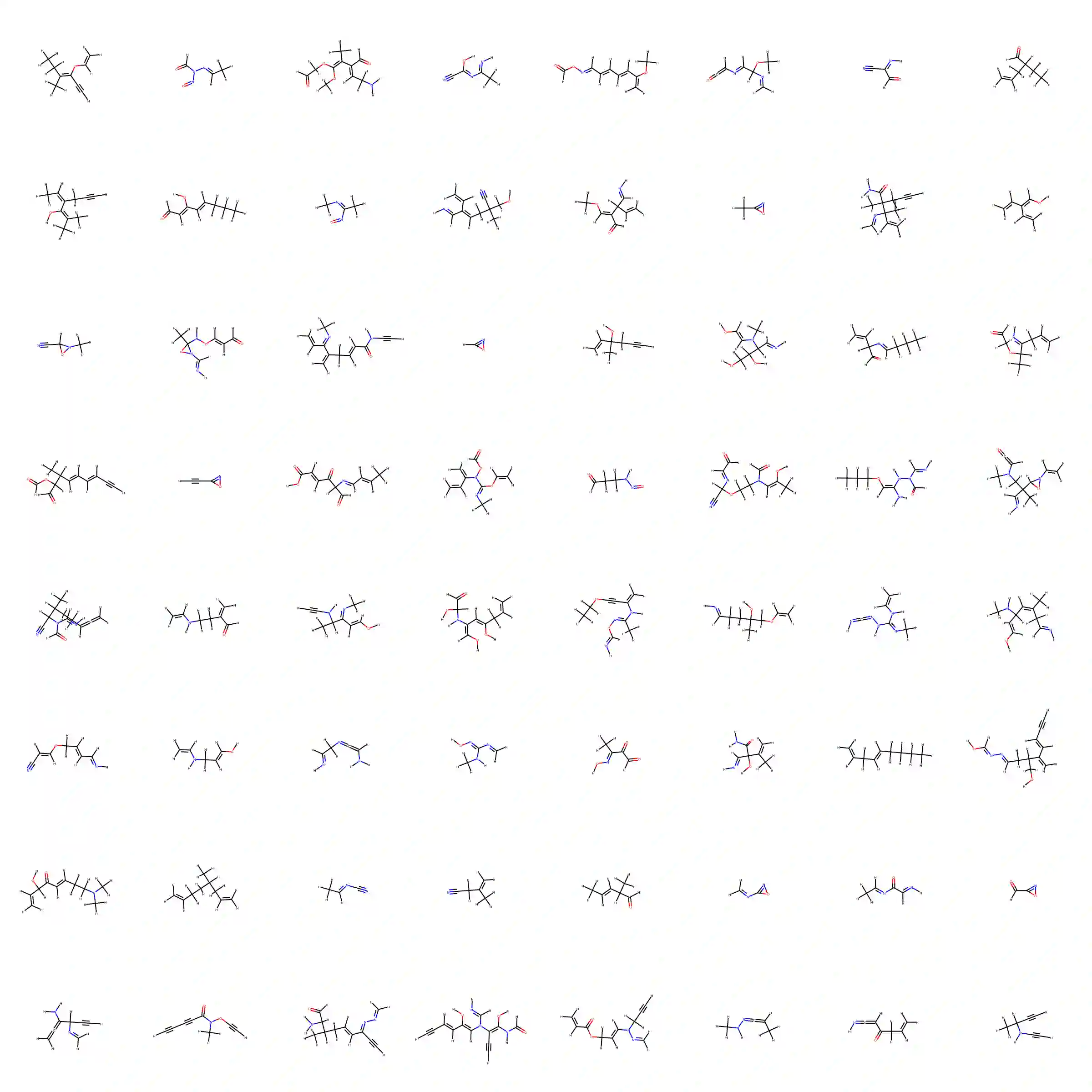
The generation of graph-structured data is an emerging problem in the field of deep learning. Various solutions have been proposed in the last few years, yet the exploration of this branch is still in an early phase. In sequential approaches, the construction of a graph is the result of a sequence of decisions, in which, at each step, a node or a group of nodes is added to the graph, along with its connections. A very relevant application of graph generation methods is the discovery of new drug molecules, which are naturally represented as graphs. In this paper, we introduce a sequential molecular graph generator based on a set of graph neural network modules, which we call MG^2N^2. Its modular architecture simplifies the training procedure, also allowing an independent retraining of a single module. The use of graph neural networks maximizes the information in input at each generative step, which consists of the subgraph produced during the previous steps. Experiments of unconditional generation on the QM9 dataset show that our model is capable of generalizing molecular patterns seen during the training phase, without overfitting. The results indicate that our method outperforms very competitive baselines, and can be placed among the state of the art approaches for unconditional generation on QM9.
翻译:图表结构数据的生成是深层学习领域出现的一个新问题。 过去几年中提出了各种解决方案, 但这一分支的探索仍处于早期阶段。 在顺序方法中, 图形的构造是一系列决定的结果, 在每个步骤中, 将节点或一组节点添加到图中, 并与其连接。 图表生成方法的一个非常相关的应用是发现新的药物分子, 这些分子自然以图表形式呈现出来。 在本文中, 我们引入了基于一组图形神经网络模块的顺序分子图形生成器, 我们称之为MG2N2。 它的模块结构简化了培训程序, 也允许对单一模块进行独立的再培训。 图形神经网络的利用使每个组合步骤的投入信息最大化, 包括前几个步骤中生成的子图。 在QM9数据集中进行无条件生成的实验表明, 我们的模型能够在培训阶段看到的一般分子模式, 而不作过度校正。 其模块结构结构简化了培训阶段, 显示, 我们的生成方法超越了具有竞争力的基准, Q9 能够将无条件的模型置于艺术生成基准和状态中。 。