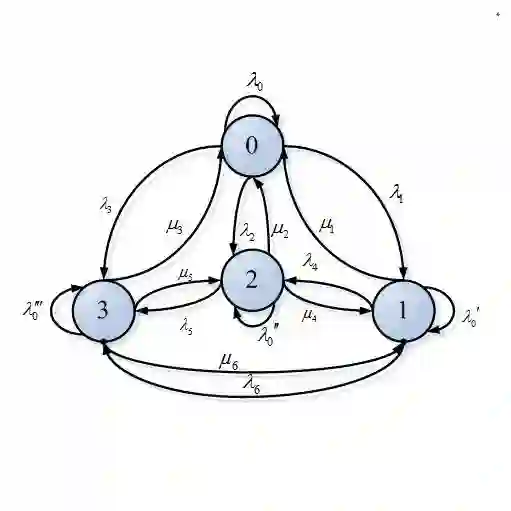
Calibrating statistical models using Bayesian inference often requires both accurate and timely estimates of parameters of interest. Particle Markov Chain Monte Carlo (p-MCMC) and Sequential Monte Carlo Squared (SMC$^2$) are two methods that use an unbiased estimate of the log-likelihood obtained from a particle filter (PF) to evaluate the target distribution. P-MCMC constructs a single Markov chain which is sequential by nature so cannot be readily parallelized using Distributed Memory (DM) architectures. This is in contrast to SMC$^2$ which includes processes, such as importance sampling, that are described as \textit{embarrassingly parallel}. However, difficulties arise when attempting to parallelize resampling. None-the-less, the choice of backward kernel, recycling scheme and compatibility with DM architectures makes SMC$^2$ an attractive option when compared with p-MCMC. In this paper, we present an SMC$^2$ framework that includes the following features: an optimal (in terms of time complexity) $\mathcal{O}(\log_2N)$ parallelization for DM architectures, an approximately optimal (in terms of accuracy) backward kernel, and an efficient recycling scheme. On a cluster of $128$ DM processors, the results on a biomedical application show that SMC$^2$ achieves up to a $70\times$ speed-up vs its sequential implementation. It is also more accurate and roughly $54\times$ faster than p-MCMC. A GitHub link is given which provides access to the code.
翻译:暂无翻译