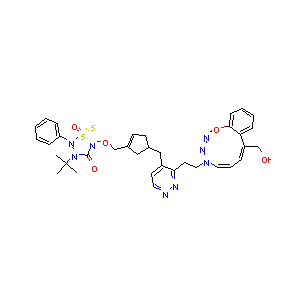
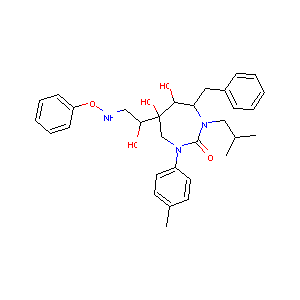
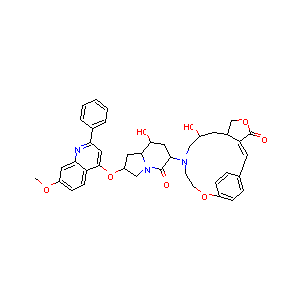
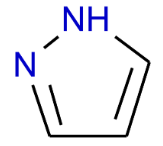
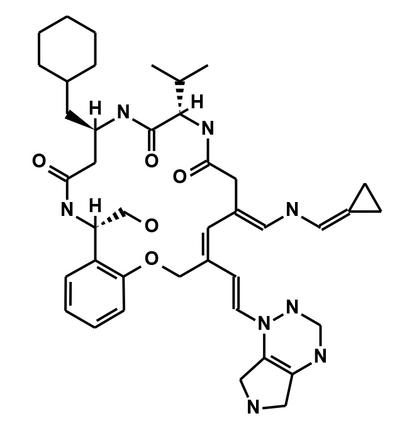
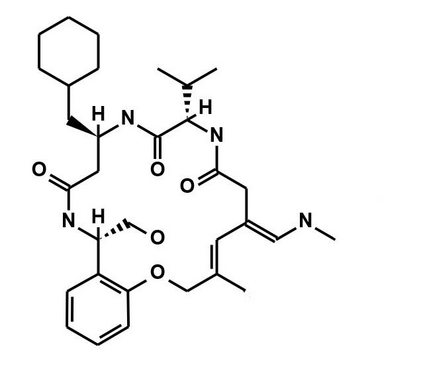
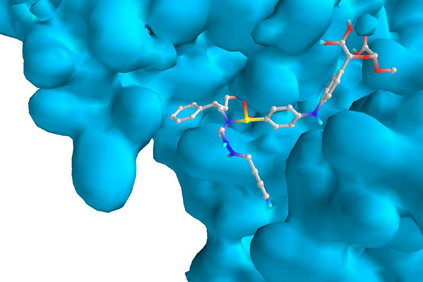
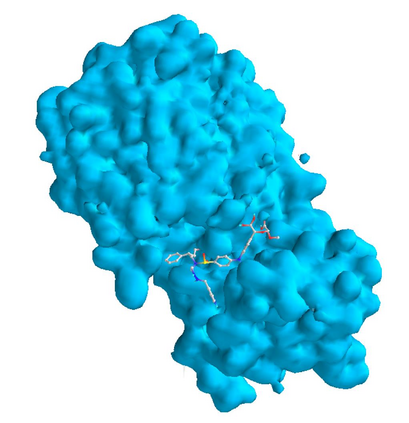
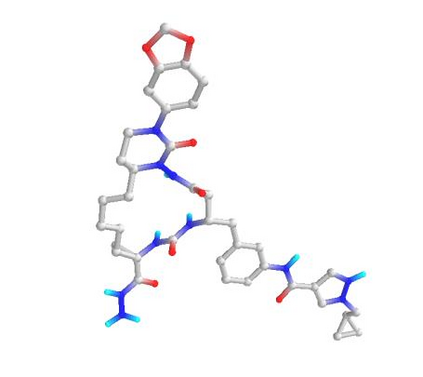
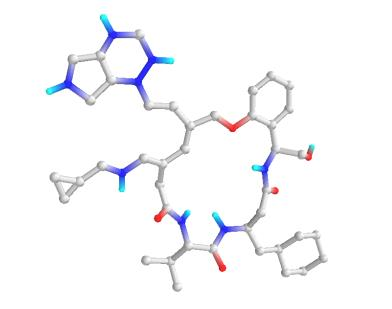
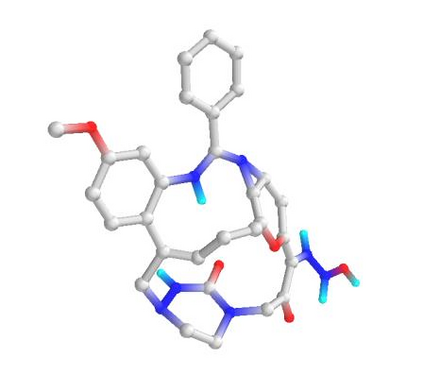
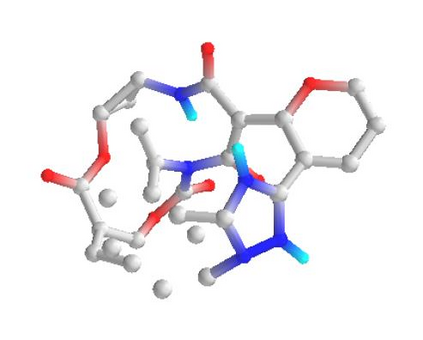
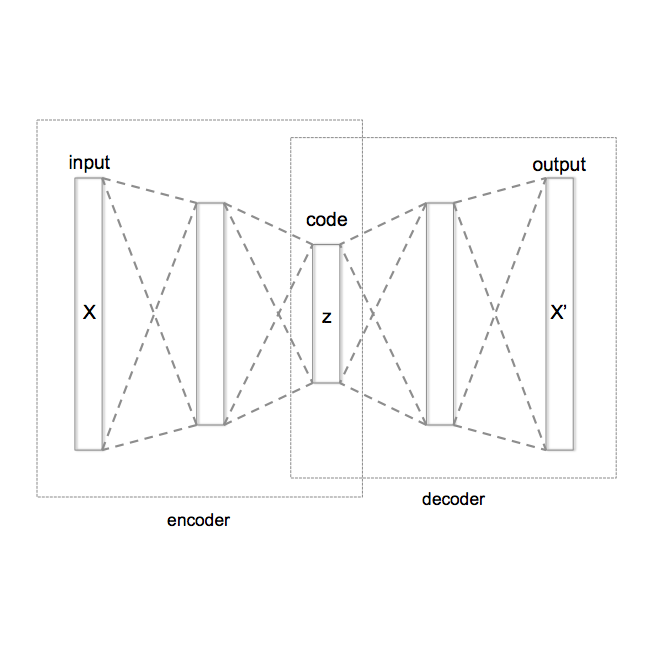
In the past several months, COVID-19 has spread over the globe and caused severe damage to the people and the society. In the context of this severe situation, an effective drug discovery method to generate potential drugs is extremely meaningful. In this paper, we provide a methodology of discovering potential drugs for the treatment of Severe Acute Respiratory Syndrome Corona-Virus 2 (commonly known as SARS-CoV-2). We proposed a new model called Genetic Constrained Graph Variational Autoencoder (GCGVAE) to solve this problem. We trained our model based on the data of various viruses' protein structure, including that of the SARS, HIV, Hep3, and MERS, and used it to generate possible drugs for SARS-CoV-2. Several optimization algorithms, including valency masking and genetic algorithm, are deployed to fine tune our model. According to the simulation, our generated molecules have great effectiveness in inhibiting SARS-CoV-2. We quantitatively calculated the scores of our generated molecules and compared it with the scores of existing drugs, and the result shows our generated molecules scores much better than those existing drugs. Moreover, our model can be also applied to generate effective drugs for treating other viruses given their protein structure, which could be used to generate drugs for future viruses.
翻译:在过去几个月里,COVID-19已经在全球蔓延,对人民和社会造成了严重损害。在这一严峻形势下,一种有效的药物发现方法产生潜在的药物是极其有意义的。在本文中,我们提供了一种方法,以发现治疗严重急性呼吸综合症科罗纳-病毒2 (俗称SARS-COV-2)的潜在药物。我们提出了一个新的模型,称为“遗传凝固图变异体2 (GCGVAE) ” 来解决这个问题。我们根据各种病毒蛋白结构的数据,包括SARS、HIV、Hep3和MERS的数据,对我们的模型进行了培训,并利用它为SARS-COV-2制造了可能的药物。一些优化算法,包括价值遮掩和基因算法,用来调整我们的模型。根据模拟,我们生成的分子在抑制SARS-COV-2(GCGVAE)方面非常有效。我们用数量计算了我们生成的分子的分数,并将其与现有药物的分数进行了比较,结果显示我们生成的分子在生产这些病毒方面比其他药物的模型还要好得多。此外,我们制作的分子可以用来生产这些病毒的病毒。