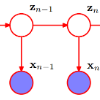
Hidden Markov models (HMMs) are probabilistic methods in which observations are seen as realizations of a latent Markov process with discrete states that switch over time. Moving beyond standard statistical tests, HMMs offer a statistical environment to optimally exploit the information present in multivariate time series, uncovering the latent dynamics that rule them. Here, we extend the Poisson HMM to the multilevel framework, accommodating variability between individuals with continuously distributed individual random effects following a lognormal distribution, and describe how to estimate the model in a fully parametric Bayesian framework. The proposed multilevel HMM enables probabilistic decoding of hidden state sequences from multivariate count time-series based on individual-specific parameters, and offers a framework to quantificate between-individual variability formally. Through a Monte Carlo study we show that the multilevel HMM outperforms the HMM for scenarios involving heterogeneity between individuals, demonstrating improved decoding accuracy and estimation performance of parameters of the emission distribution, and performs equally well when not between heterogeneity is present. Finally, we illustrate how to use our model to explore the latent dynamics governing complex multivariate count data in an empirical application concerning pilot whale diving behaviour in the wild, and how to identify neural states from multi-electrode recordings of motor neural cortex activity in a macaque monkey in an experimental set up. We make the multilevel HMM introduced in this study publicly available in the R-package mHMMbayes in CRAN.
翻译:暂无翻译