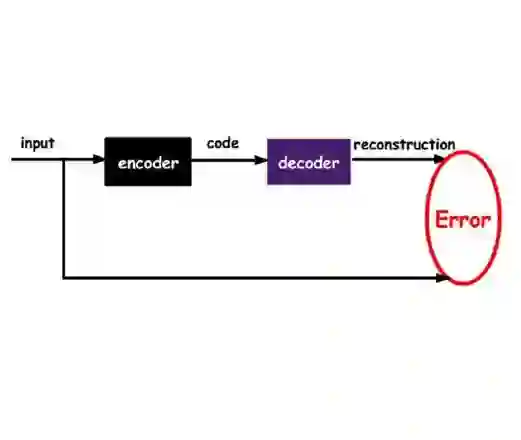
Cancer is a complex disease with significant social and economic impact. Advancements in high-throughput molecular assays and the reduced cost for performing high-quality multi-omics measurements have fuelled insights through machine learning . Previous studies have shown promise on using multiple omic layers to predict survival and stratify cancer patients. In this paper, we developed a Supervised Autoencoder (SAE) model for survival-based multi-omic integration which improves upon previous work, and report a Concrete Supervised Autoencoder model (CSAE), which uses feature selection to jointly reconstruct the input features as well as predict survival. Our experiments show that our models outperform or are on par with some of the most commonly used baselines, while either providing a better survival separation (SAE) or being more interpretable (CSAE). We also perform a feature selection stability analysis on our models and notice that there is a power-law relationship with features which are commonly associated with survival. The code for this project is available at: https://github.com/phcavelar/coxae
翻译:癌症是一种复杂的疾病,具有重大的社会和经济影响。高通量分子分析的进步以及高质量多工程测量成本的降低,通过机器学习激发了深刻的洞察力 。以前的研究表明,使用多种显微层预测存活率和将癌症病人分解为有希望。在本文中,我们开发了一种以生存为基础的多工程集成的受监督自动编码器(SAE)模型,该模型改进了以前的工作,并报告了一个具体受监督的自动编码模型(CSAE),该模型利用特征选择来联合重建输入特征并预测生存率。我们的实验显示,我们的模型的性能超过或与最常用的一些基线相当,同时提供更好的生存分离(SAE)或更易于解释(CSAE)。我们还对我们的模型进行特征选择稳定性分析,并通知我们与通常与生存有关的特征存在权力-法律关系。该项目的代码见:https://github.com/phcavelar/cexue。