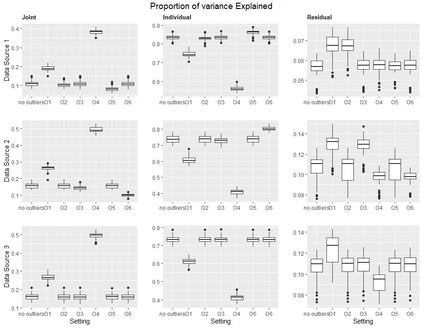
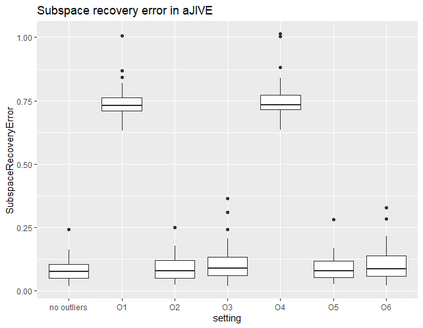
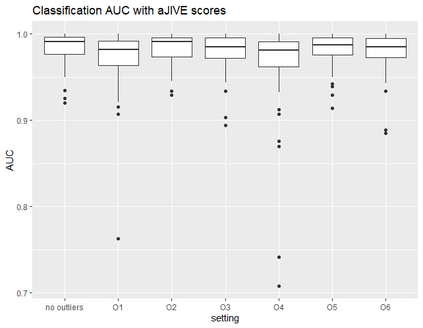
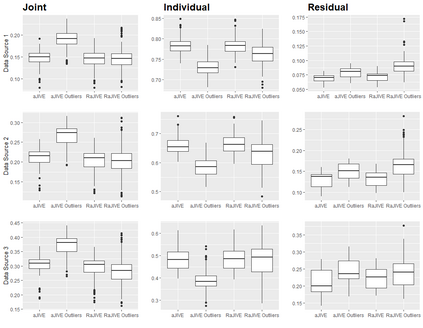
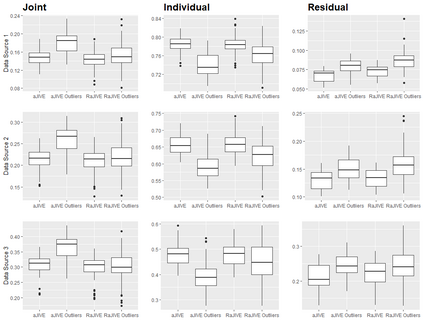
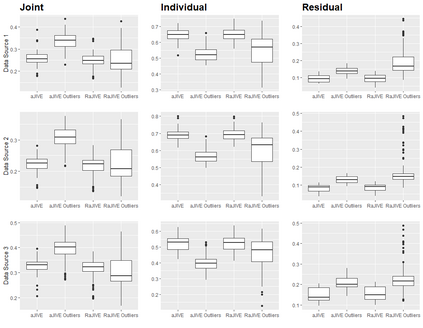
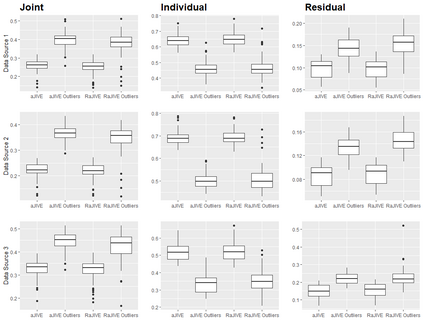
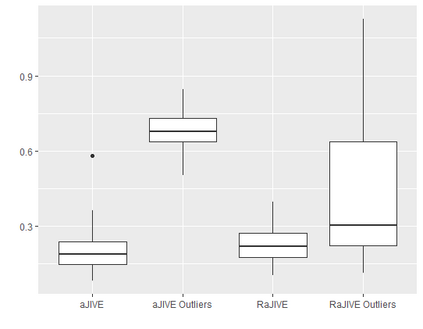
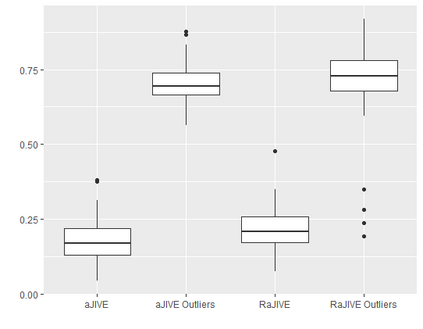
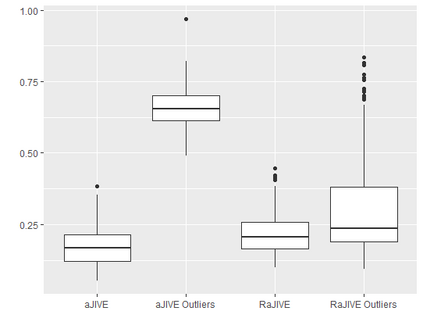
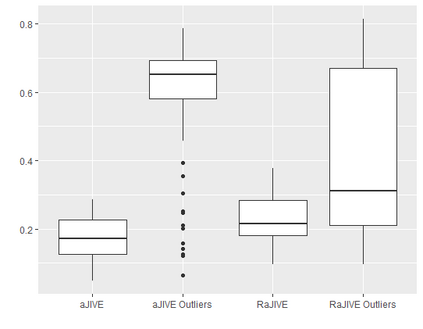
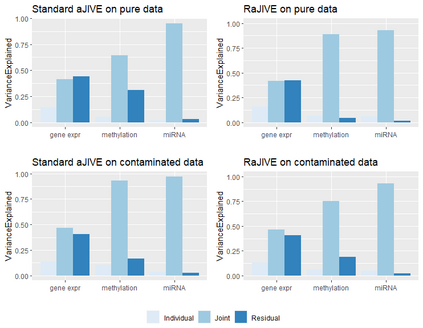
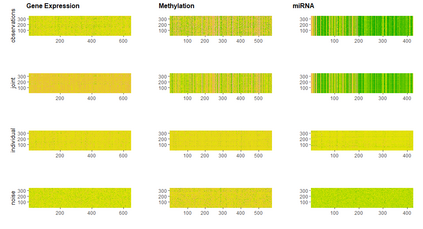
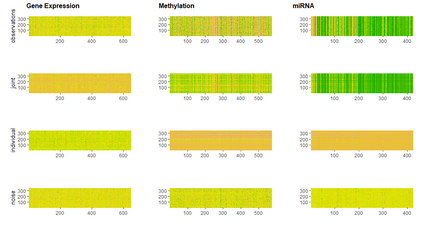
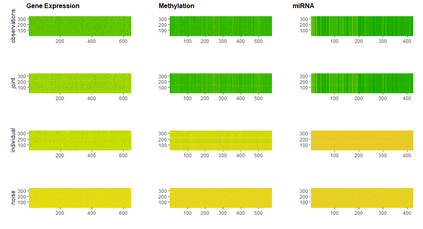
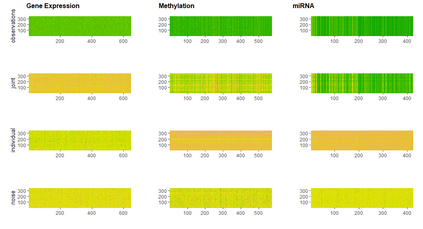
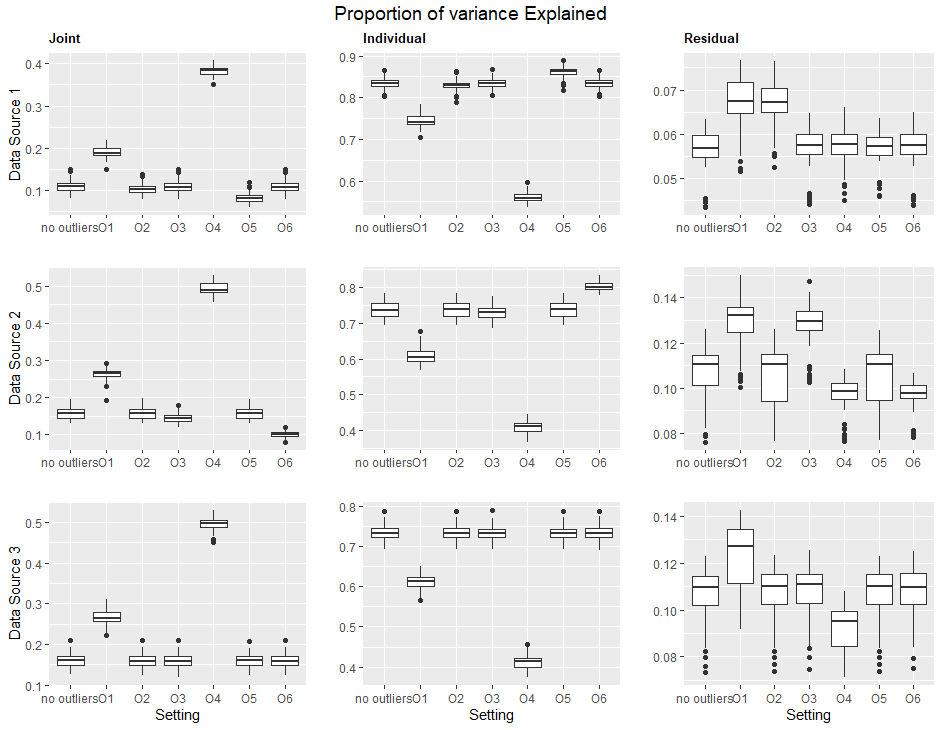
With increasing availability of high dimensional, multi-source data, the identification of joint and data specific patterns of variability has become a subject of interest in many research areas. Several matrix decomposition methods have been formulated for this purpose, for example JIVE (Joint and Individual Variation Explained), and its angle based variation, aJIVE. Although the effect of data contamination on the estimated joint and individual components has not been considered in the literature, gross errors and outliers in the data can cause instability in such methods, and lead to incorrect estimation of joint and individual variance components. We focus on the aJIVE factorization method and provide a thorough analysis of the effect outliers on the resulting variation decomposition. After showing that such effect is not negligible when all data-sources are contaminated, we propose a robust extension of aJIVE (RaJIVE) that integrates a robust formulation of the singular value decomposition into the aJIVE approach. The proposed RaJIVE is shown to provide correct decompositions even in the presence of outliers and improves the performance of aJIVE. We use extensive simulation studies with different levels of data contamination to compare the two methods. Finally, we describe an application of RaJIVE to a multi-omics breast cancer dataset from The Cancer Genome Atlas. We provide the R package RaJIVE with a ready-to-use implementation of the methods and documentation of code and examples.
翻译:随着高维、多源数据的日益可得性,确定联合和数据具体变异模式已成为许多研究领域关注的一个主题,为此制定了若干矩阵分解方法,例如JIVE(联合和个人变异解释)及其基于角度的变异。虽然文献中未考虑到数据污染对估计的联合和个别组成部分的影响,但数据中的重大错误和偏差可能导致这类方法的不稳定,并导致对联合和个别差异组成部分的不正确估计。我们注重准因子化方法,对由此产生的变异变异变异变异变异的影响外端进行透彻分析。在显示所有数据源受到污染时,这种影响并非微不足道之后,我们建议大力扩展AJIVE(拉维),将单值分解变的可靠配制纳入JIVE方法。拟议的RJIVE系统显示,即使存在异端,也能够提供正确的分解分解结果,并改进JVE的性能。我们使用广泛的模拟研究,用不同程度的数据模型来比较RIVI系统实施方法。我们用不同程度的RIVI系统模型来比较两种方法。
相关内容
- Today (iOS and OS X): widgets for the Today view of Notification Center
- Share (iOS and OS X): post content to web services or share content with others
- Actions (iOS and OS X): app extensions to view or manipulate inside another app
- Photo Editing (iOS): edit a photo or video in Apple's Photos app with extensions from a third-party apps
- Finder Sync (OS X): remote file storage in the Finder with support for Finder content annotation
- Storage Provider (iOS): an interface between files inside an app and other apps on a user's device
- Custom Keyboard (iOS): system-wide alternative keyboards
Source: iOS 8 Extensions: Apple’s Plan for a Powerful App Ecosystem