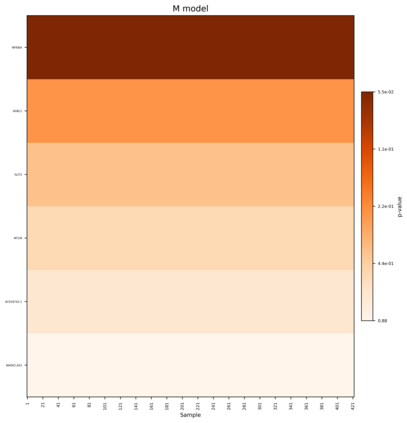
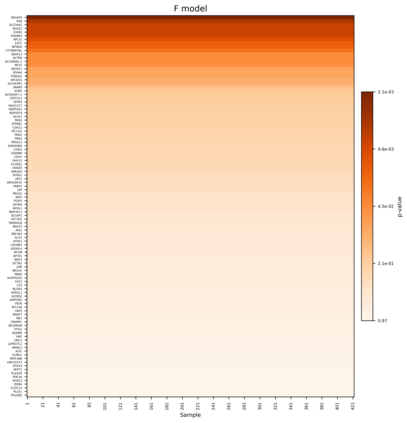
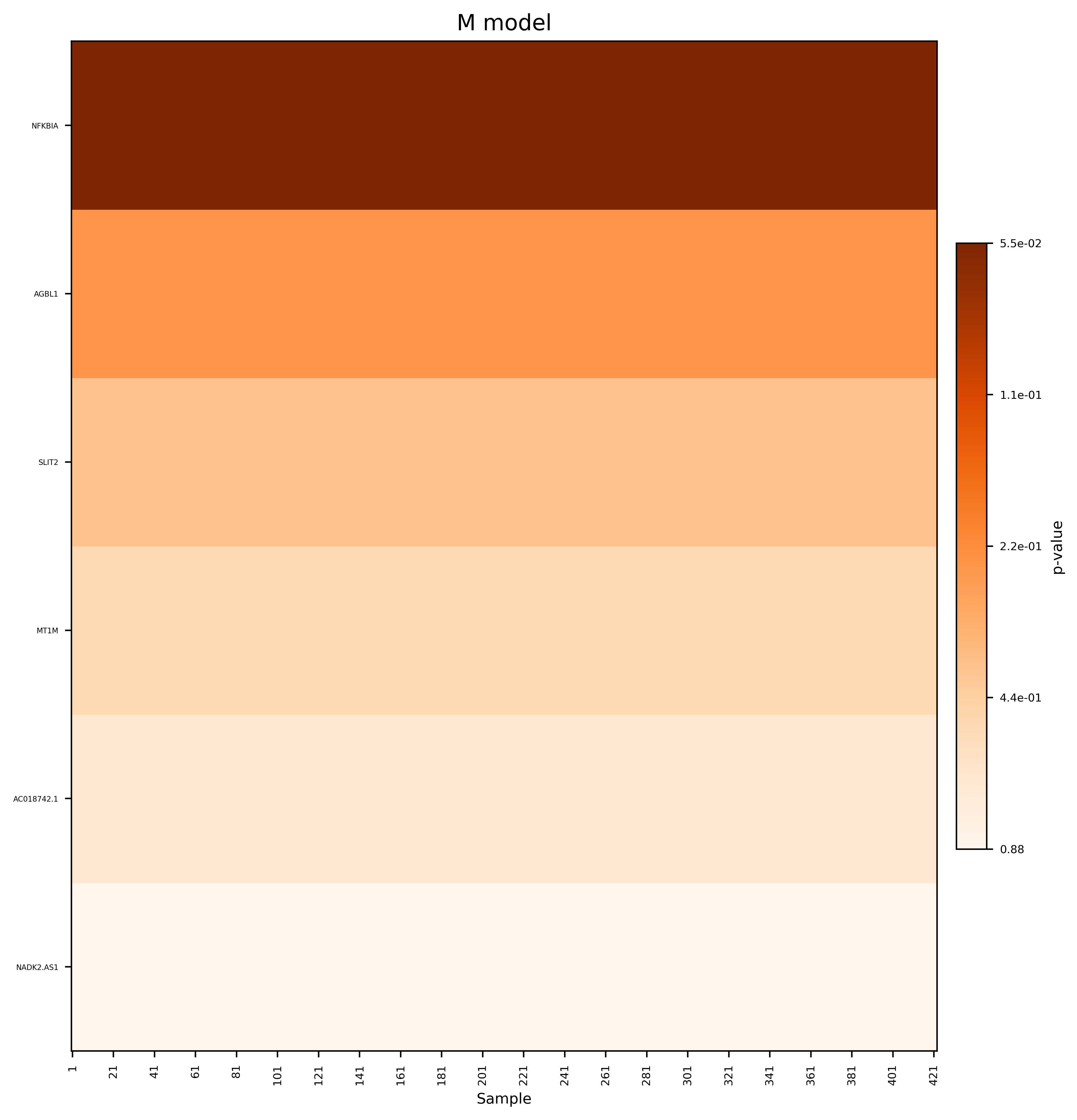
Single-cell RNA sequencing (scRNA-seq) has revolutionized the study of cellular heterogeneity, enabling detailed molecular profiling at the individual cell level. However, integrating high-dimensional single-cell data into causal mediation analysis remains challenging due to zero inflation and complex mediator structures. We propose a novel mediation framework leveraging zero-inflated negative binomial models to characterize cell-level mediator distributions and beta regression for zero-inflation proportions. The model can identify expression level as well as expressed proportion that could mediate disease-leading causal pathway. Extensive simulation studies demonstrate improved power and controlled false discovery rates. We further illustrate the utility of this approach through application to ROSMAP single-cell transcriptomic data, uncovering biologically meaningful mediation effects that enhance understanding of disease mechanisms.
翻译:暂无翻译