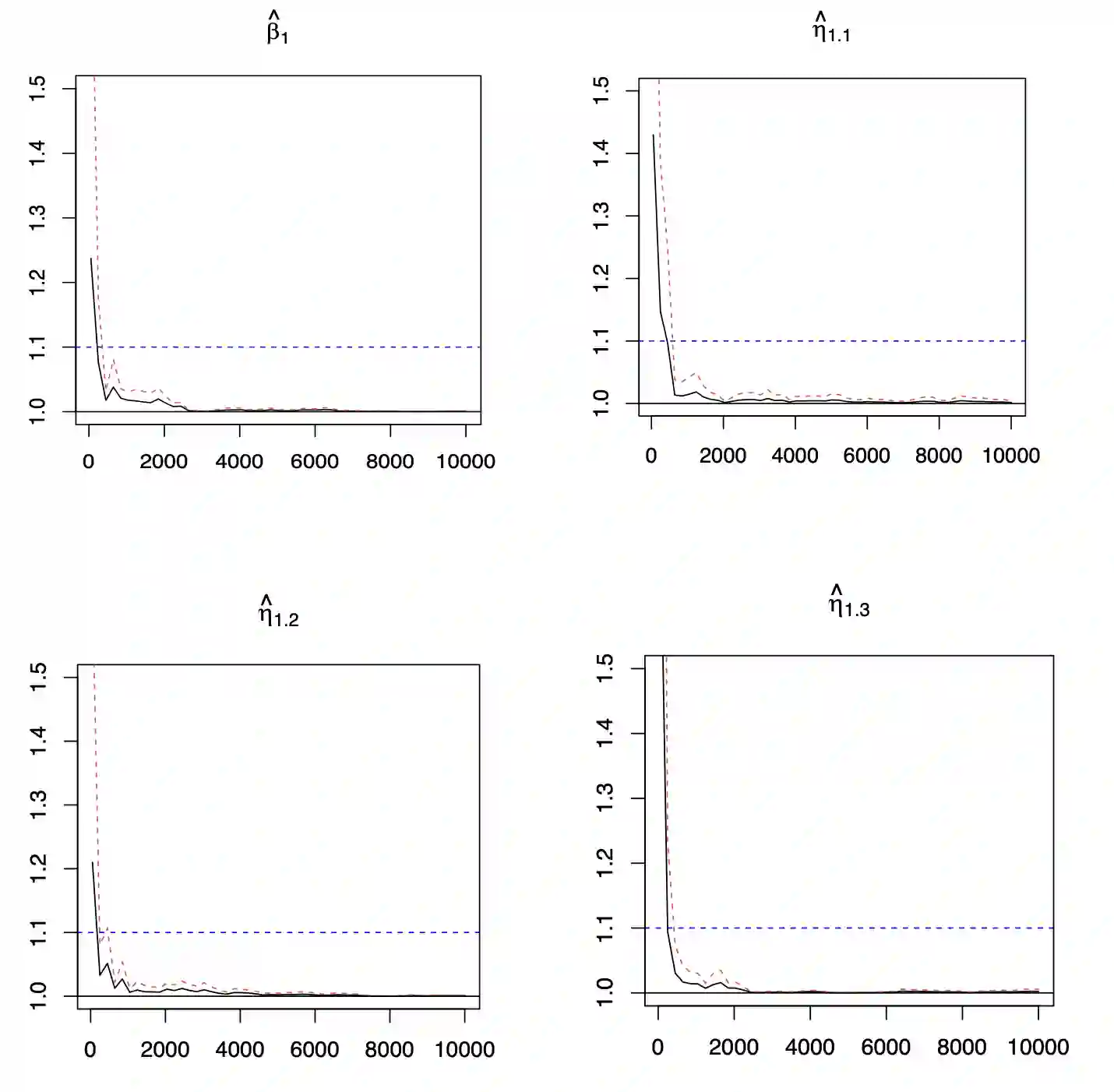
In high-throughput genetics studies, an important aim is to identify gene-environment interactions associated with the clinical outcomes. Recently, multiple marginal penalization methods have been developed and shown to be effective in G$\times$E studies. However, within the Bayesian framework, marginal variable selection has not received much attention. In this study, we propose a novel marginal Bayesian variable selection method for G$\times$E studies. In particular, our marginal Bayesian method is robust to data contamination and outliers in the outcome variables. With the incorporation of spike-and-slab priors, we have implemented the Gibbs sampler based on MCMC. The proposed method outperforms a number of alternatives in extensive simulation studies. The utility of the marginal robust Bayesian variable selection method has been further demonstrated in the case studies using data from the Nurse Health Study (NHS). Some of the identified main and interaction effects from the real data analysis have important biological implications.
翻译:在高通量遗传学研究中,一个重要目标是确定与临床结果有关的基因-环境相互作用。最近,开发了多种边际惩罚方法,并在G$\times$E研究中证明这些方法行之有效。然而,在巴伊西亚框架内,边际变量选择没有受到多少关注。在本研究中,我们建议为G$\times $E研究采用新的边际巴伊西亚变量选择方法。特别是,我们的边际巴伊西亚方法对数据污染和结果变量的外部效应十分有力。随着加附了钉子和碎片前缀,我们采用了以MCMC为基础的Gibs取样器。拟议方法在广泛的模拟研究中优于若干替代方法。边际强大的巴伊斯变量选择方法在利用护士健康研究(NHS)数据进行的案例研究中得到了进一步证明。一些查明的主要影响和相互作用对实际数据分析具有重要的生物影响。