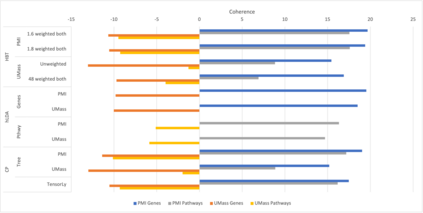
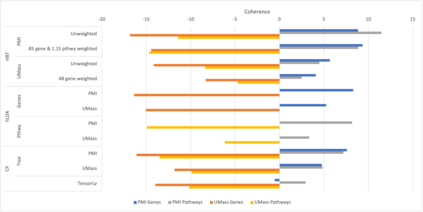
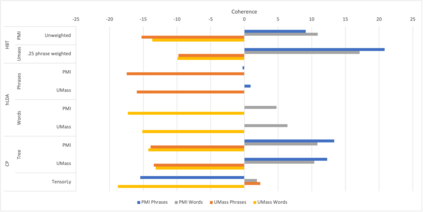
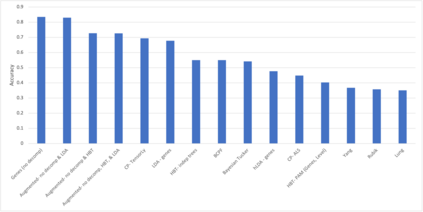
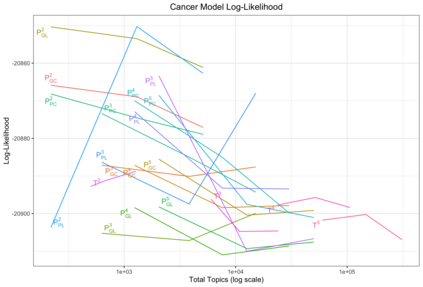
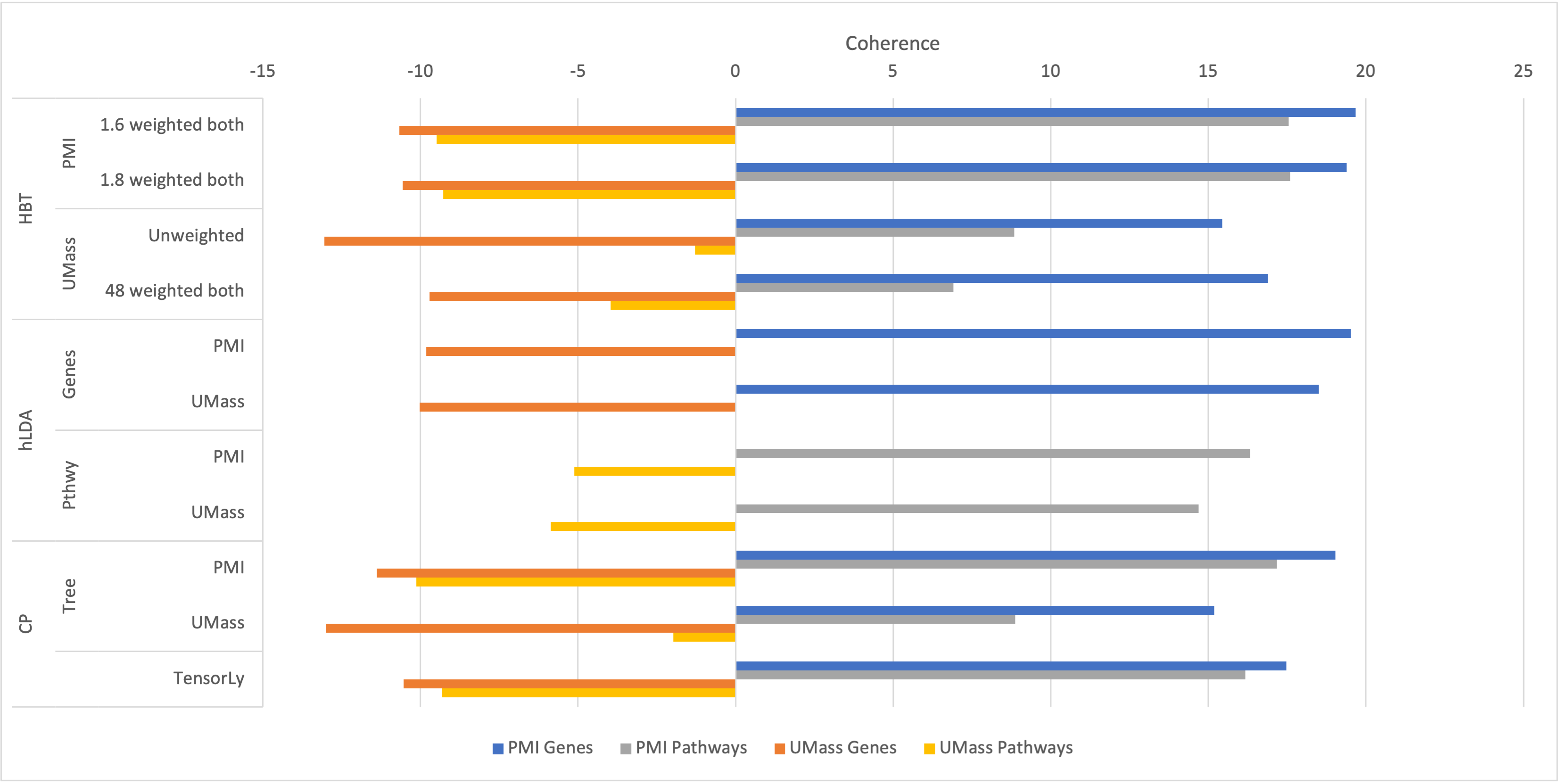
We develop methods for reducing the dimensionality of large data sets, common in biomedical applications. Learning about patients using genetic data often includes more features than observations, which makes direct supervised learning difficult. One method of reducing the feature space is to use latent Dirichlet allocation to group genetic variants in an unsupervised manner. Latent Dirichlet allocation describes a patient as a mixture of topics corresponding to genetic variants. This can be generalized as a Bayesian tensor decomposition to account for multiple feature variables. Our most significant contributions are with hierarchical topic modeling. We design distinct methods of incorporating hierarchical topic modeling, based on nested Chinese restaurant processes and Pachinko Allocation Machine, into Bayesian tensor decomposition. We apply these models to examine patients with one of four common types of cancer (breast, lung, prostate, and colorectal) and siblings with and without autism spectrum disorder. We linked the genes with their biological pathways and combine this information into a tensor of patients, counts of their genetic variants, and the genes' membership in pathways. We find that our trained models outperform baseline models, with respect to coherence, by up to 40%.
翻译:生物医学应用中常见的、关于使用基因数据的病人的学习往往包括更多的特征,而不是观测,这使得直接监督的学习困难。减少特征空间的方法之一是以不受监督的方式将潜在的迪里赫特分配用于组群遗传变异。 中迪里赫特分配将病人描述为与基因变异相对应的混合课题。 这可以被广泛称为一种巴伊西亚的强力分解,以考虑到多种特征变量。 我们最重要的贡献是等级主题模型。 我们设计了将等级主题模型纳入Bayesian Exwards 和 Pachinko 分配机器的不同方法。 我们用这些模型来检查四种常见癌症(乳癌、肺癌、前列腺癌和彩色切除症)之一的病人和患有自闭症的兄弟姐妹。 我们将这些基因与其生物路径连接起来,并将这一信息与病人的气压、基因变数和路径中的基因基因组合结合起来。 我们发现,我们经过训练的模型超越了基准模型的模型,以40 %的一致性。