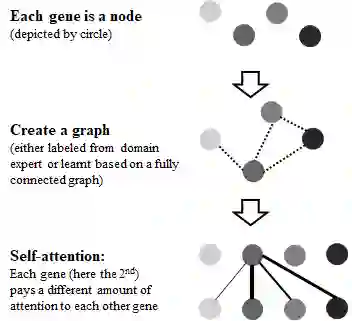
Genetic mutations can cause disease by disrupting normal gene function. Identifying the disease-causing mutations from millions of genetic variants within an individual patient is a challenging problem. Computational methods which can prioritize disease-causing mutations have, therefore, enormous applications. It is well-known that genes function through a complex regulatory network. However, existing variant effect prediction models only consider a variant in isolation. In contrast, we propose VEGN, which models variant effect prediction using a graph neural network (GNN) that operates on a heterogeneous graph with genes and variants. The graph is created by assigning variants to genes and connecting genes with an gene-gene interaction network. In this context, we explore an approach where a gene-gene graph is given and another where VEGN learns the gene-gene graph and therefore operates both on given and learnt edges. The graph neural network is trained to aggregate information between genes, and between genes and variants. Variants can exchange information via the genes they connect to. This approach improves the performance of existing state-of-the-art models.
翻译:基因突变可以通过破坏正常基因功能而导致疾病。 确定个人病人中数百万种基因变异导致的疾病变异是一个具有挑战性的问题。 能够优先考虑疾病变异的计算方法因此具有巨大的应用性。 众所周知,基因通过复杂的监管网络发挥作用。 然而, 现有的变异效应预测模型只能孤立地考虑一个变异。 相反, 我们提议VEGN, 使用一个带有基因和变异的图形神经网络( GNN) 运行的图形神经网络( GNN) 模型影响预测。 该图是通过给基因分配变异和将基因与基因- 基因互动网络连接起来的方法创建的。 在这方面, 我们探索了一种方法, 即提供基因基因图, 另一种方法是VEGN 学习基因基因图, 并在给定的边缘和学到的边缘进行操作。 图形神经网络经过培训, 将基因之间以及基因和变异种之间的信息汇总起来。 变种可以通过它们连接的基因交换信息。 这种方法可以改进现有状态模型的性能。