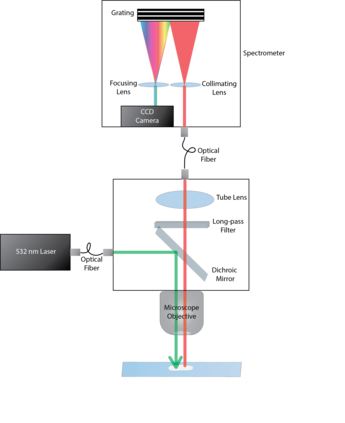
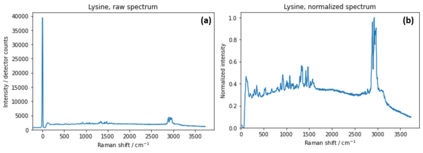
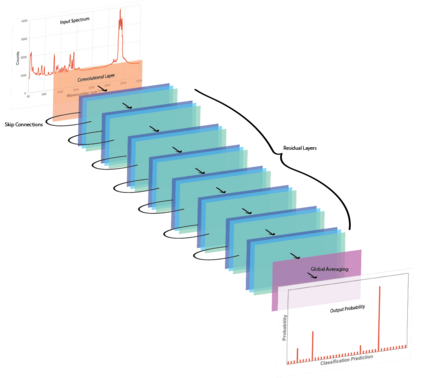
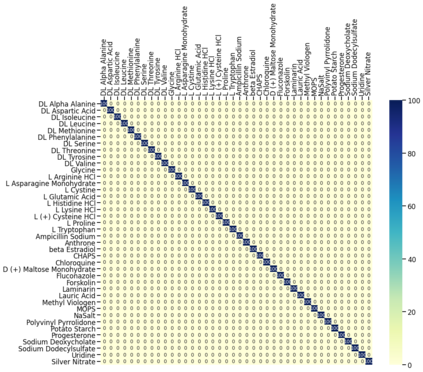
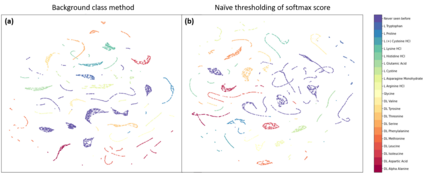
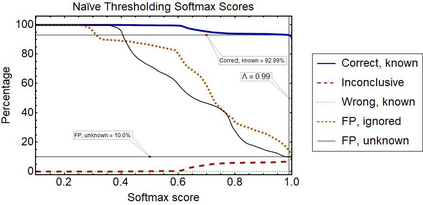
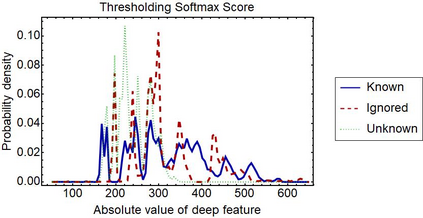
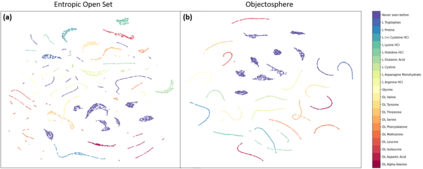
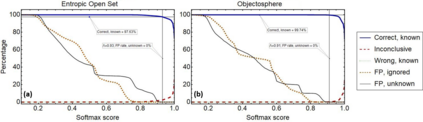
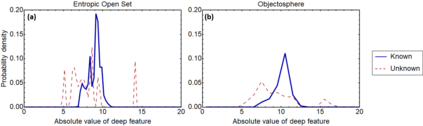
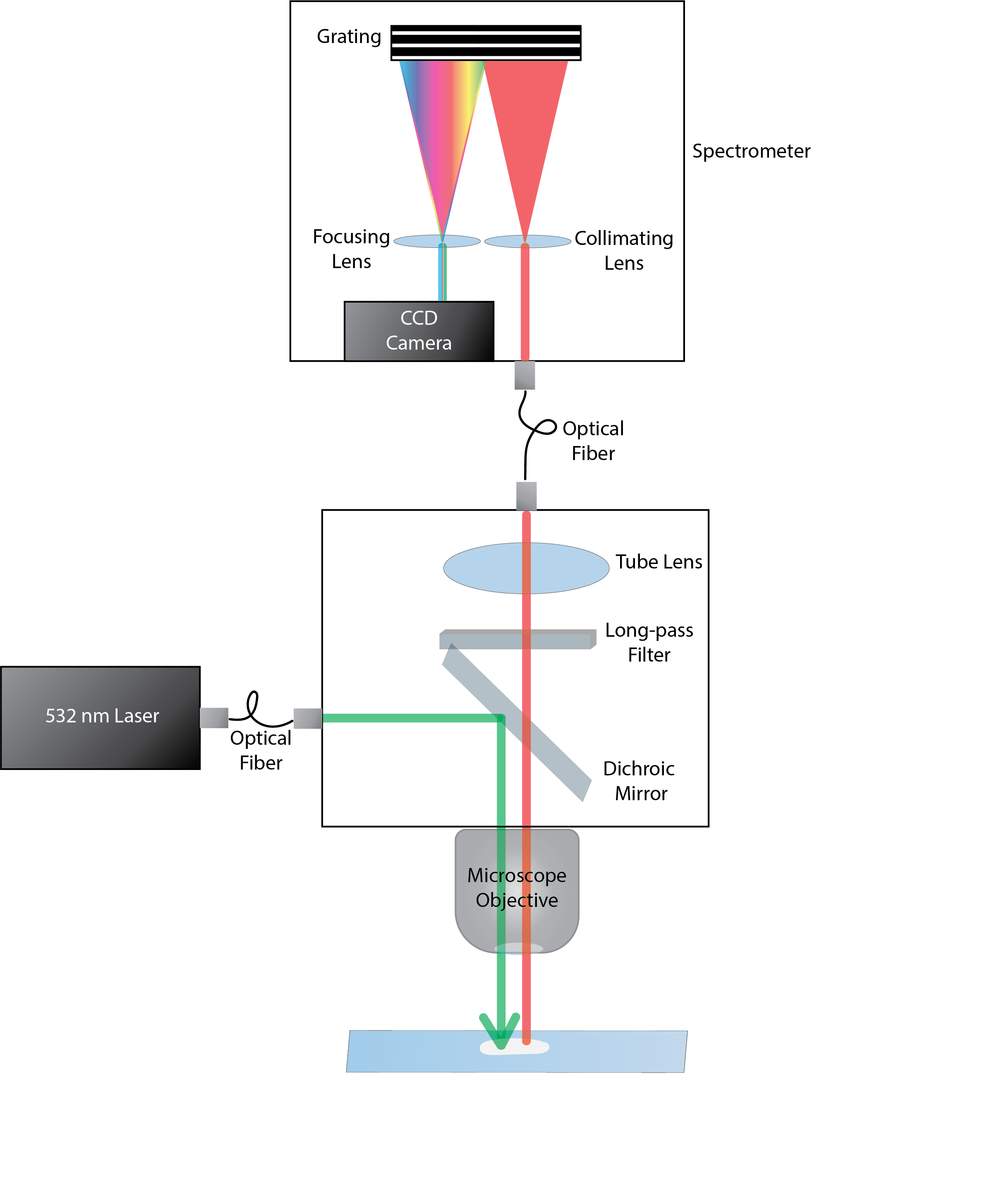
Raman spectroscopy in combination with machine learning has significant promise for applications in clinical settings as a rapid, sensitive, and label-free identification method. These approaches perform well in classifying data that contains classes that occur during the training phase. However, in practice, there are always substances whose spectra have not yet been taken or are not yet known and when the input data are far from the training set and include new classes that were not seen at the training stage, a significant number of false positives are recorded which limits the clinical relevance of these algorithms. Here we show that these obstacles can be overcome by implementing recently introduced Entropic Open Set and Objectosphere loss functions. To demonstrate the efficiency of this approach, we compiled a database of Raman spectra of 40 chemical classes separating them into 20 biologically relevant classes comprised of amino acids, 10 irrelevant classes comprised of bio-related chemicals, and 10 classes that the Neural Network has not seen before, comprised of a variety of other chemicals. We show that this approach enables the network to effectively identify the unknown classes while preserving high accuracy on the known ones, dramatically reducing the number of false positives while preserving high accuracy on the known classes, which will allow this technique to bridge the gap between laboratory experiments and clinical applications.
翻译:与机器学习相结合的拉曼光谱和机器光谱分析在临床应用中作为快速、敏感和无标签的识别方法有很大的希望。这些方法在对包含培训阶段发生的各类数据的数据进行分类方面表现良好。然而,在实践中,总是有一些物质,其光谱尚未被采纳,或尚不为人知,输入数据远离培训组,包括培训阶段未见的新课程,记录了大量假正数,限制了这些算法的临床相关性。我们在这里表明,通过实施最近引入的 Entropic Open Set 和 对象层损失功能,这些障碍是可以克服的。为了展示这一方法的效率,我们汇编了一个由40个化学类组成的拉曼光谱数据库,将它们分为20个生物相关类别,包括氨酸,10个由生物相关化学品组成的相关课程,以及Neural网络以前未曾见过的10个课程,由各种化学品组成。我们表明,这一方法使网络能够有效地识别未知的类别,同时保持已知的高度精确性,极大地减少假正值数量,同时保持已知的临床实验应用之间的高度差距。