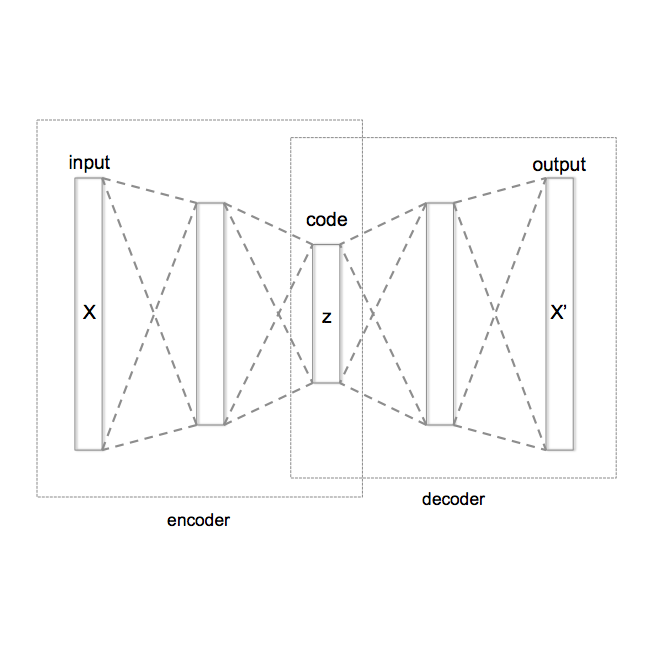
For personalized medicines, very crucial intrinsic information is present in high dimensional omics data which is difficult to capture due to the large number of molecular features and small number of available samples. Different types of omics data show various aspects of samples. Integration and analysis of multi-omics data give us a broad view of tumours, which can improve clinical decision making. Omics data, mainly DNA methylation and gene expression profiles are usually high dimensional data with a lot of molecular features. In recent years, variational autoencoders (VAE) have been extensively used in embedding image and text data into lower dimensional latent spaces. In our project, we extend the idea of using a VAE model for low dimensional latent space extraction with the self-supervised learning technique of feature subsetting. With VAEs, the key idea is to make the model learn meaningful representations from different types of omics data, which could then be used for downstream tasks such as cancer type classification. The main goals are to overcome the curse of dimensionality and integrate methylation and expression data to combine information about different aspects of same tissue samples, and hopefully extract biologically relevant features. Our extension involves training encoder and decoder to reconstruct the data from just a subset of it. By doing this, we force the model to encode most important information in the latent representation. We also added an identity to the subsets so that the model knows which subset is being fed into it during training and testing. We experimented with our approach and found that SubOmiEmbed produces comparable results to the baseline OmiEmbed with a much smaller network and by using just a subset of the data. This work can be improved to integrate mutation-based genomic data as well.
翻译:对于个性化药物来说,由于大量分子特征和少量现有样本,难以获取的高维显性数据中存在非常关键的内在信息。不同种类的显性数据显示样本的各个方面。多缩性数据的整合和分析使我们对肿瘤有了广泛的了解,从而可以改善临床决策。对基因数据,主要是DNA甲基化和基因表达剖面,通常是具有大量分子特征的高维数据。近年来,变异自动变异基因组(VAE)被广泛用于将图像和文本数据嵌入低维性潜伏空间。在我们的项目中,我们扩展了使用低维隐性空间提取的VAE模型以及自我超强的特征子集化技术的构想。对于VAE来说,关键的想法是让模型从不同种类的奥米基数据中学习有意义的描述,然后用于下游任务,例如癌症类型分类。主要目标是克服维度的诅咒,将甲基和表达数据整合成关于低维度潜基层潜值的图像数据。我们希望,我们用这个深度的模型和生物基集层数据 将我们用来做一个基础化的模型,然后将一个基础数据提取一个基础数据,然后从一个基础数据,然后从我们的组织中找到一个基础数据。
相关内容
- Today (iOS and OS X): widgets for the Today view of Notification Center
- Share (iOS and OS X): post content to web services or share content with others
- Actions (iOS and OS X): app extensions to view or manipulate inside another app
- Photo Editing (iOS): edit a photo or video in Apple's Photos app with extensions from a third-party apps
- Finder Sync (OS X): remote file storage in the Finder with support for Finder content annotation
- Storage Provider (iOS): an interface between files inside an app and other apps on a user's device
- Custom Keyboard (iOS): system-wide alternative keyboards
Source: iOS 8 Extensions: Apple’s Plan for a Powerful App Ecosystem