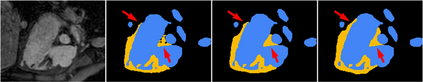
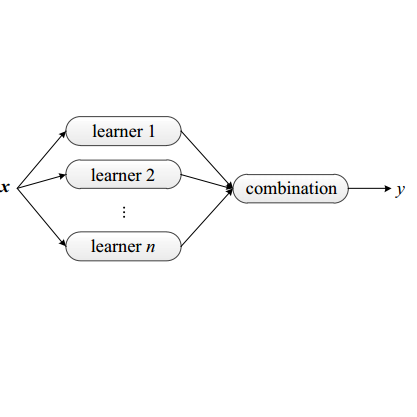
3D image segmentation plays an important role in biomedical image analysis. Many 2D and 3D deep learning models have achieved state-of-the-art segmentation performance on 3D biomedical image datasets. Yet, 2D and 3D models have their own strengths and weaknesses, and by unifying them together, one may be able to achieve more accurate results. In this paper, we propose a new ensemble learning framework for 3D biomedical image segmentation that combines the merits of 2D and 3D models. First, we develop a fully convolutional network based meta-learner to learn how to improve the results from 2D and 3D models (base-learners). Then, to minimize over-fitting for our sophisticated meta-learner, we devise a new training method that uses the results of the base-learners as multiple versions of "ground truths". Furthermore, since our new meta-learner training scheme does not depend on manual annotation, it can utilize abundant unlabeled 3D image data to further improve the model. Extensive experiments on two public datasets (the HVSMR 2016 Challenge dataset and the mouse piriform cortex dataset) show that our approach is effective under fully-supervised, semi-supervised, and transductive settings, and attains superior performance over state-of-the-art image segmentation methods.
翻译:3D 图像分割在生物医学图像分析中起着重要作用。 许多 2D 和 3D 深层学习模型在 3D 生物医学图像数据集中取得了最先进的分解性表现。 然而, 2D 和 3D 模型有其自身的优势和弱点,并且通过将它们统一起来,也许能够取得更准确的结果。 在本文件中,我们为3D 生物医学图像分割提出了一个新的混合学习框架,将 2D 和 3D 模型的优点结合起来。 首先,我们开发了一个基于 3D 和 3D 模型的完全连锁网络, 来学习如何改进 2D 和 3D 模型( 基线利纳 ) 的结果。 然后, 为了尽可能减少我们尖端的元脱盲模型的过度匹配, 我们设计了一个新的培训方法, 将基- leaner 的结果作为“ 地面真相” 的多种版本。 此外, 由于我们新的 3D- Lea 培训计划并不取决于手动的注释, 它可以利用大量的未贴标签的 3D 图像数据数据数据来进一步改进模型。 在两种公共数据集上进行广泛的实验( HVStrevial- sqrevial- droviduformax ) 。