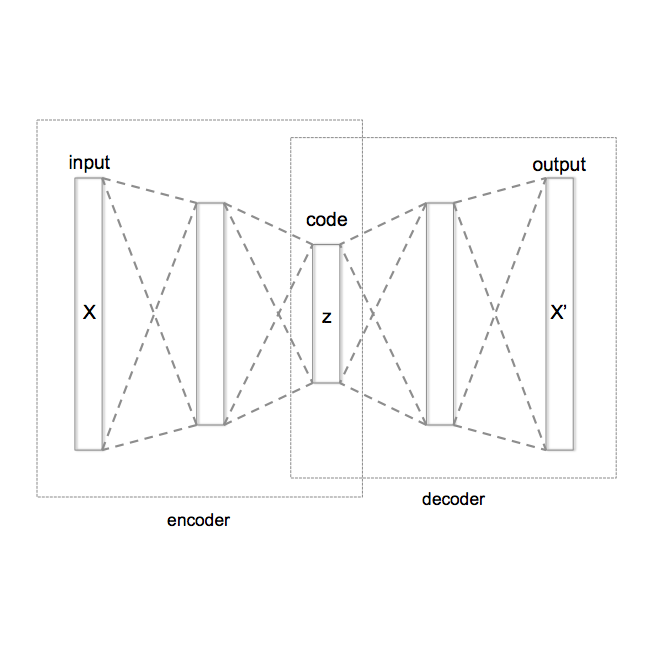
Deep generative models have emerged as a powerful tool for learning informative molecular representations and designing novel molecules with desired properties, with applications in drug discovery and material design. Deep generative auto-encoders defined over molecular SMILES strings have been a popular choice for that purpose. However, capturing salient molecular properties like quantum-chemical energies remains challenging and requires sophisticated neural net models of molecular graphs or geometry-based information. As a simpler and more efficient alternative, we present a SMILES Variational Auto-Encoder (VAE) augmented with topological data analysis (TDA) representations of molecules, known as persistence images. Our experiments show that this TDA augmentation enables a SMILES VAE to capture the complex relation between 3D geometry and electronic properties, and allows generation of novel, diverse, and valid molecules with geometric features consistent with the training data, which exhibit a varying range of global electronic structural properties, such as a small HOMO-LUMO gap - a critical property for designing organic solar cells. We demonstrate that our TDA augmentation yields better success in downstream tasks compared to models trained without these representations and can assist in targeted molecule discovery.
翻译:深基因模型已成为学习信息分子表达方式和设计具有理想特性的新分子的有力工具,在药物发现和材料设计中应用了这种应用。在分子SMILES字符串上定义的深基因自动编码器是这方面的一种流行选择。然而,捕捉量化学能源等突出分子特性仍然具有挑战性,需要分子图或基于几何的信息的尖端神经网模型。作为一个更简单、更有效率的替代方法,我们提出了一个SMILES变形自动计算机(VAE),它与被称为持久性图像的分子的表层数据分析(TDA)相补充。我们的实验显示,这种扩增使SMILESVAE能够捕捉到3D几何特性与电子特性之间的复杂关系,并允许产生与培训数据相一致的具有几何特征的新颖、多样化和有效分子。这种模型展示了各种全球电子结构特性,例如小型的HOMO-LUMO差距,这是设计有机太阳能电池的关键特性。我们证明,我们的TDA增强作用使下游任务与没有这些表象的模型相比更成功,能够有针对性地发现。