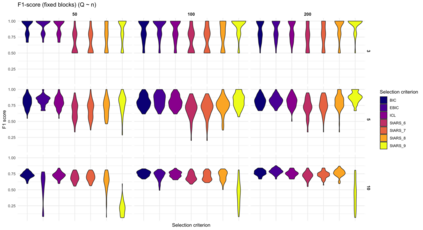
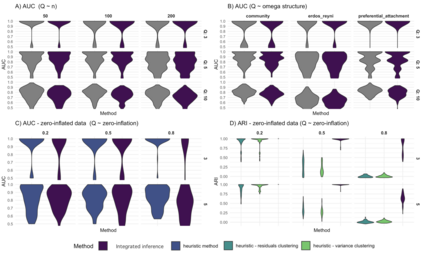
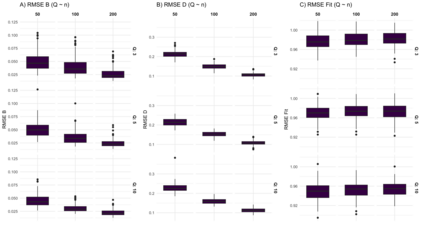
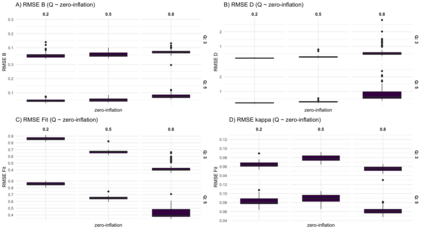
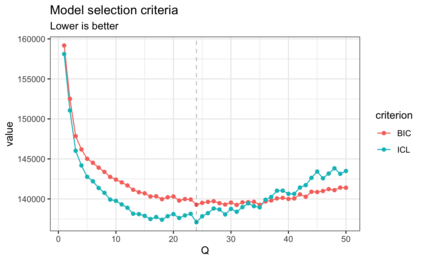
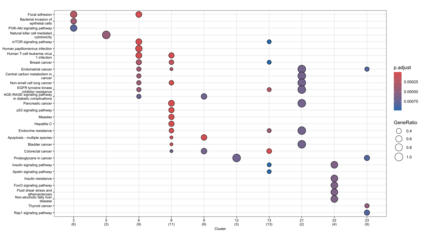
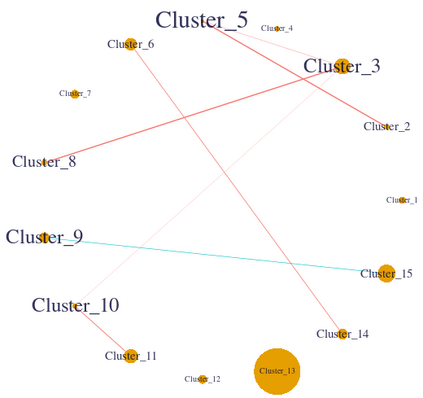
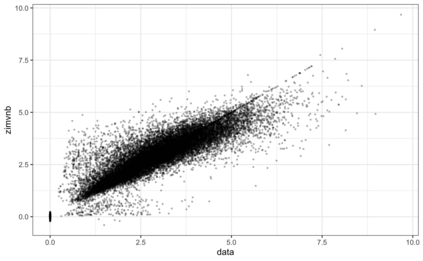
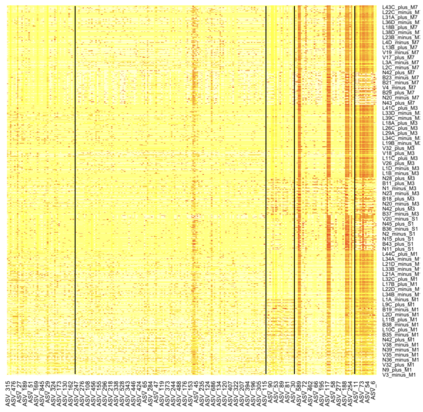
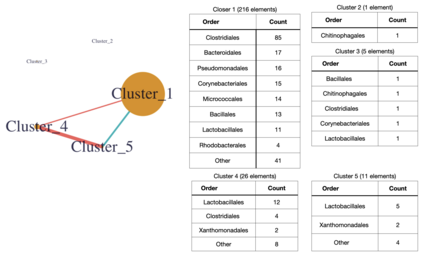
We consider high dimensional Gaussian graphical models inference. These models provide a rigorous framework to describe a network of statistical dependencies between entities, such as genes in genomic regulation studies or species in ecology. Penalized methods, including the standard Graphical-Lasso, are well-known approaches to infer the parameters of these models. As the number of variables in the model (of entities in the network) grow, the network inference and interpretation become more complex. We propose Normal-Block, a new model that clusters variables and consider a network at the cluster level. Normal-Block both adds structure to the network and reduces its size. We build on Graphical-Lasso to add a penalty on the network's edges and limit the detection of spurious dependencies, we also propose a zero-inflated version of the model to account for real-world data properties. For the inference procedure, we propose a direct heuristic method and another more rigorous one that simultaneously infers the clustering of variables and the association network between clusters, using a penalized variational Expectation-Maximization approach. An implementation of the model in R, in a package called normalblockr, is available on github (https://github.com/jeannetous/normalblockr). We present the results in terms of clustering and network inference using both simulated data and various types of real-world data (proteomics, words occurrences on webpages, and microbiota distribution).
翻译:暂无翻译