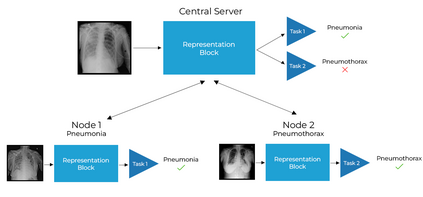
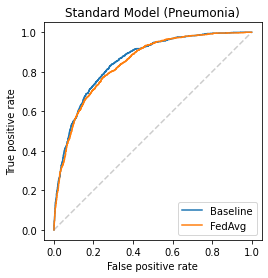
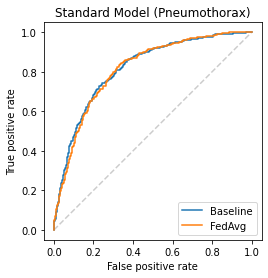
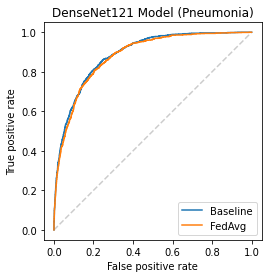
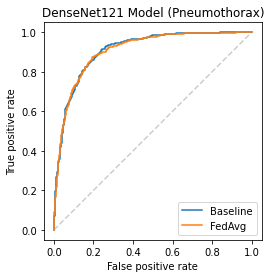
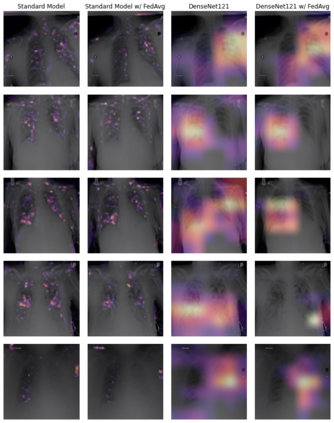
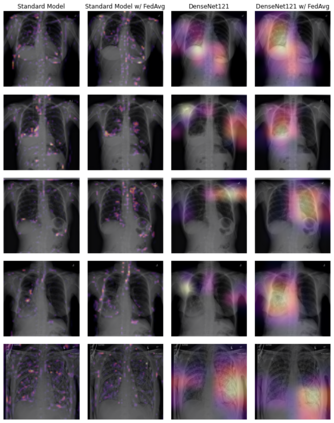
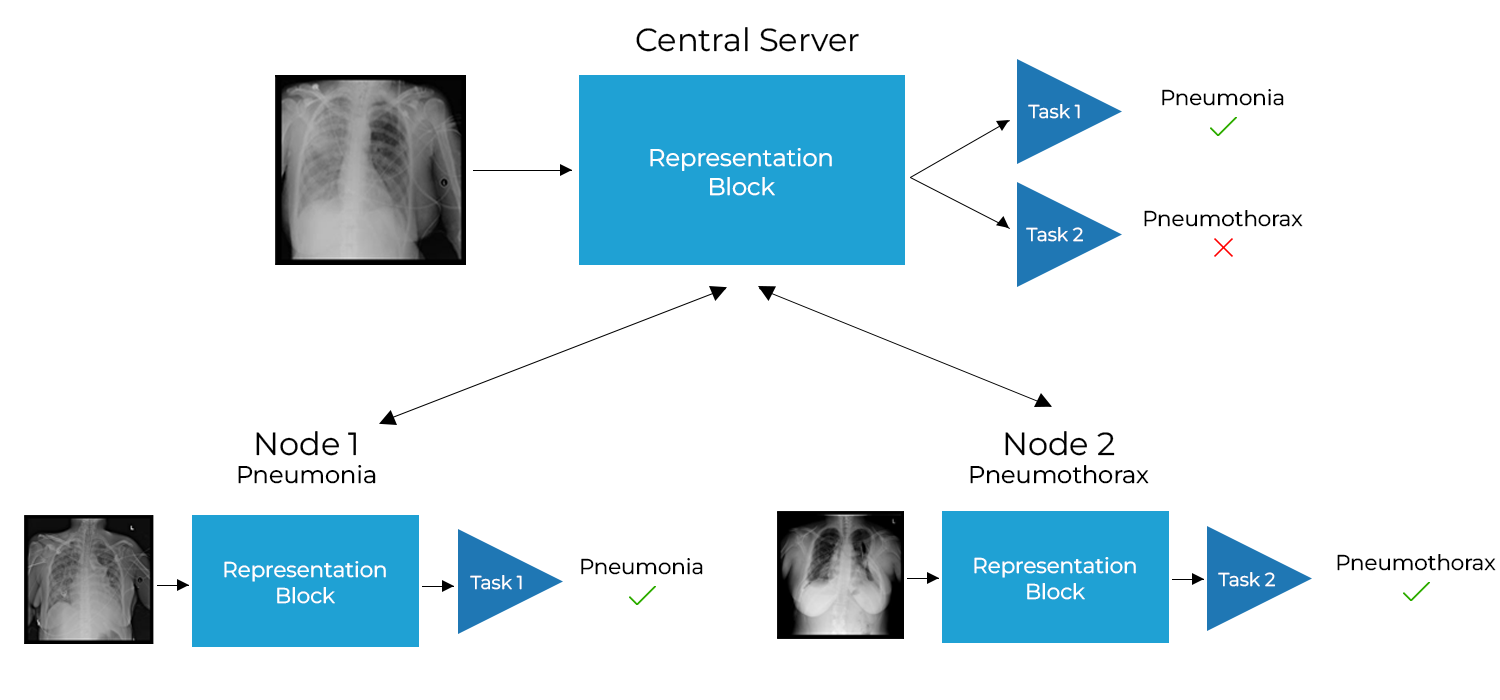
Chest X-ray (CXR) datasets hosted on Kaggle, though useful from a data science competition standpoint, have limited utility in clinical use because of their narrow focus on diagnosing one specific disease. In real-world clinical use, multiple diseases need to be considered since they can co-exist in the same patient. In this work, we demonstrate how federated learning (FL) can be used to make these toy CXR datasets from Kaggle clinically useful. Specifically, we train a single FL classification model (`global`) using two separate CXR datasets -- one annotated for presence of pneumonia and the other for presence of pneumothorax (two common and life-threatening conditions) -- capable of diagnosing both. We compare the performance of the global FL model with models trained separately on both datasets (`baseline`) for two different model architectures. On a standard, naive 3-layer CNN architecture, the global FL model achieved AUROC of 0.84 and 0.81 for pneumonia and pneumothorax, respectively, compared to 0.85 and 0.82, respectively, for both baseline models (p>0.05). Similarly, on a pretrained DenseNet121 architecture, the global FL model achieved AUROC of 0.88 and 0.91 for pneumonia and pneumothorax, respectively, compared to 0.89 and 0.91, respectively, for both baseline models (p>0.05). Our results suggest that FL can be used to create global `meta` models to make toy datasets from Kaggle clinically useful, a step forward towards bridging the gap from bench to bedside.
翻译:在卡格格勒(Kaggle)托管的切斯特X射线(CXR)数据集,虽然从数据科学竞争的角度来说是有用的,但在临床使用方面用处有限,因为它们狭隘地侧重于诊断一种特定疾病。在现实世界临床使用中,需要考虑多种疾病,因为它们在同一病人中同时存在。在这项工作中,我们展示了如何利用由卡格格勒(Kagggle)托管的玩具CXR数据集。具体地说,我们用两个单独的CXR数据集来培训单一的FL分类模型(`Global'),使用两个单独的CXR数据集 -- -- 一个是肺炎的存在附加说明的,另一个是肺炎(两种常见和危及生命的条件)的存在。在现实世界临床使用中,多个疾病模型(FLL)的性能与在两个不同的模型上分别培训过的模型(`基准线'FLL')的性能进行比较。在标准、天真的3级CNN架构中,全球FL模型从0.84和0.81的基数差距从0.81到肺炎和肺炎和肺部阵列阵列的基数(分别从0.80比、0.8至0.8和0.8的FNS的基数),在基准中分别是ANS的模型,在0.80至0.8至0.8和0.8至0.8和0.8和0.8至0.8和0.8和0.8的模型的模型。