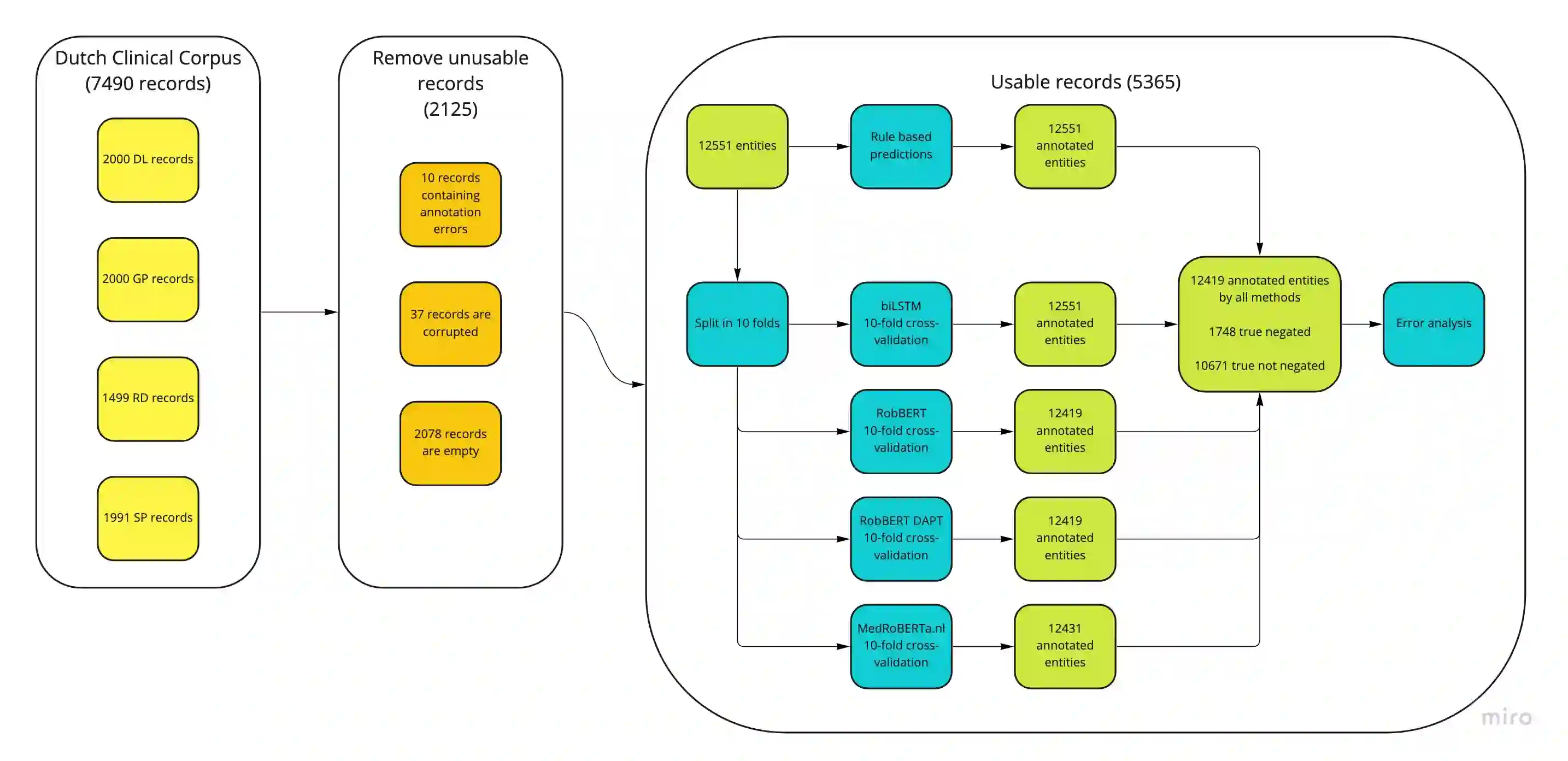
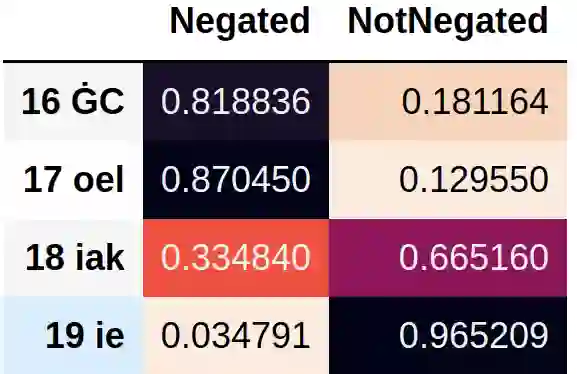
As structured data are often insufficient, labels need to be extracted from free text in electronic health records when developing models for clinical information retrieval and decision support systems. One of the most important contextual properties in clinical text is negation, which indicates the absence of findings. We aimed to improve large scale extraction of labels by comparing three methods for negation detection in Dutch clinical notes. We used the Erasmus Medical Center Dutch Clinical Corpus to compare a rule-based method based on ContextD, a biLSTM model using MedCAT and (finetuned) RoBERTa-based models. We found that both the biLSTM and RoBERTa models consistently outperform the rule-based model in terms of F1 score, precision and recall. In addition, we systematically categorized the classification errors for each model, which can be used to further improve model performance in particular applications. Combining the three models naively was not beneficial in terms of performance. We conclude that the biLSTM and RoBERTa-based models in particular are highly accurate accurate in detecting clinical negations, but that ultimately all three approaches can be viable depending on the use case at hand.
翻译:由于结构化数据往往不足,在开发临床信息检索和决定支持系统模型时,需要从电子健康记录中的免费文本中提取标签。临床文本中最重要的背景特性之一是否定,这表明没有结果。我们的目标是通过比较荷兰临床说明中的三种否定检测方法来改进大规模提取标签;我们利用Erasmus医疗中心荷兰临床公司比较基于环境D的基于规则的方法,一种使用医疗医疗治疗方法的BILSTM模型和(经改进的)RoBERTA模型。我们发现,BILSTM和RoBERTA模型在F1评分、精确度和回顾方面始终超越基于规则的模型。此外,我们系统地分类了每一种模型的分类错误,可以用来进一步改进模型在具体应用中的性能。我们天真地将这三种模型结合在性能方面是无益的。我们的结论是,基于BILSTM和RoBERTA的模型在检测临床否定方面特别准确,但最终所有三种方法都能够根据手用案例而可行。