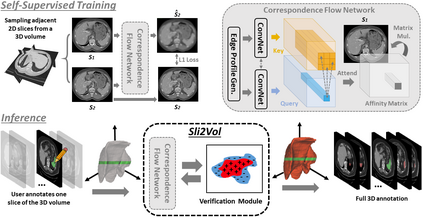
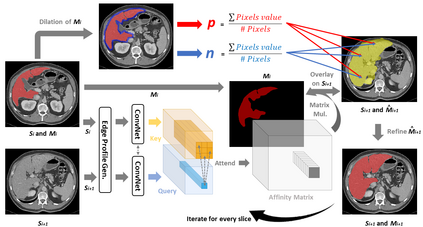
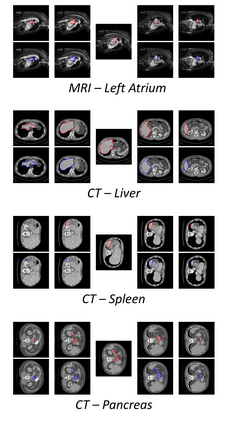
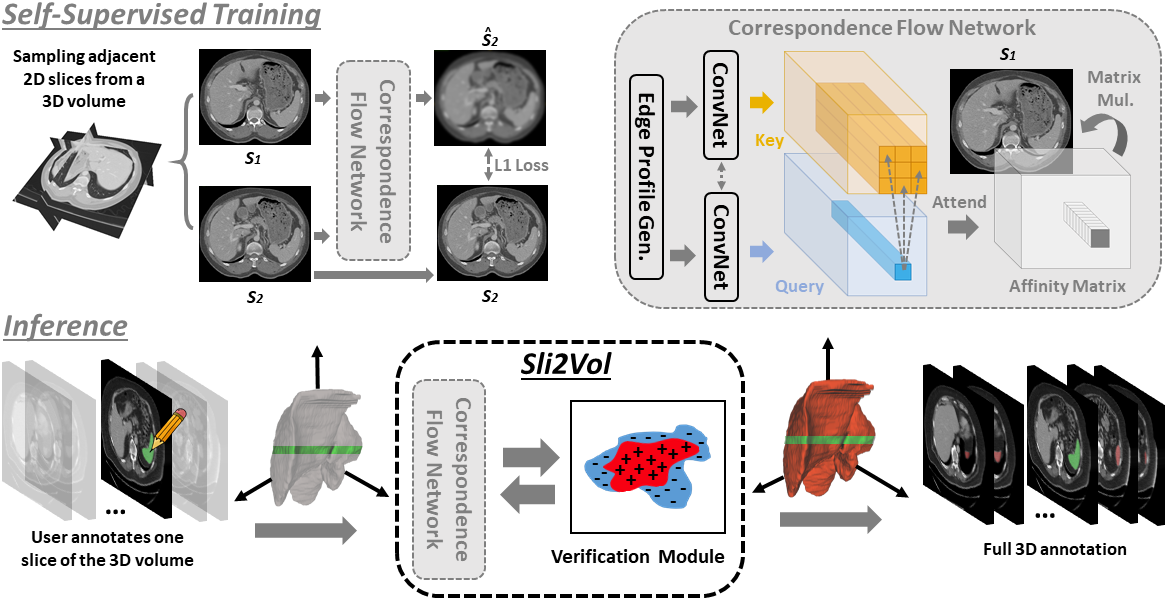
The objective of this work is to segment any arbitrary structures of interest (SOI) in 3D volumes by only annotating a single slice, (i.e. semi-automatic 3D segmentation). We show that high accuracy can be achieved by simply propagating the 2D slice segmentation with an affinity matrix between consecutive slices, which can be learnt in a self-supervised manner, namely slice reconstruction. Specifically, we compare the proposed framework, termed as Sli2Vol, with supervised approaches and two other unsupervised/ self-supervised slice registration approaches, on 8 public datasets (both CT and MRI scans), spanning 9 different SOIs. Without any parameter-tuning, the same model achieves superior performance with Dice scores (0-100 scale) of over 80 for most of the benchmarks, including the ones that are unseen during training. Our results show generalizability of the proposed approach across data from different machines and with different SOIs: a major use case of semi-automatic segmentation methods where fully supervised approaches would normally struggle. The source code will be made publicly available at https://github.com/pakheiyeung/Sli2Vol.
翻译:这项工作的目标是将3D卷中的任何任意利益结构(SOI)分解为3D卷中的任何任意利益结构(SOI),只说明一个片段(即半自动3D分割)。我们表明,只要在连续切片之间传播2D切片分割,同时在连续切片之间以亲监督的方式学习,即切片重建,就可以实现高度的精确性。具体地说,我们比较了称为Sli2Vol的拟议框架,有监督的方法和另外两种不受监督/自监督的切片登记方法,涉及8个公共数据集(CT和MRI扫描),横跨9个不同的SOIs。在没有任何参数调整的情况下,该模型在大多数基准中,包括在培训期间看不见的基准中,以Dice分数(0-100级)达到优异性性性表现。我们的结果显示,拟议方法在来自不同机器的数据和不同的SOIS:一个主要使用半自动分离方法,通常在完全监督的方法中挣扎。源代码将在https://githliub.com/pakheung上公开提供。