We introduce Permutation and Structured Perturbation Inference (PSPI), a new problem formulation that abstracts many graph matching tasks that arise in systems biology. PSPI can be viewed as a robust formulation of the permutation inference or graph matching, where the objective is to find a permutation between two graphs under the assumption that a set of edges may have undergone a perturbation due to an underlying cause. For example, suppose there are two gene regulatory networks X and Y from a diseased and normal tissue respectively. Then, the PSPI problem can be used to detect if there has been a structural change between the two networks which can serve as a signature of the disease. Besides the new problem formulation, we propose an ADMM algorithm (STEPD) to solve a relaxed version of the PSPI problem. An extensive case study on comparative gene regulatory networks (GRNs) is used to demonstrate that STEPD is able to accurately infer structured perturbations and thus provides a tool for computational biologists to identify novel prognostic signatures. A spectral analysis confirms that STEPD can recover small clique-like perturbations making it a useful tool for detecting permutation-invariant changes in graphs.
翻译:我们引入了变形和结构性扰动推断(PSPI),这是一种新问题配方,可以摘要归纳系统生物学中出现的许多图表匹配任务。PSPI可以被视为对变形推断或图形匹配的有力配方,目的是在假设一系列边缘可能因根本原因而经历扰动的情况下,在两个图表之间找到一种变异。例如,假设一个疾病和正常组织分别有两个基因调节网络X和Y。然后,PSPI问题可用于检测这两个网络之间是否发生了结构性变化,这些变化可以作为疾病的特征。除了新的问题配方外,我们建议采用ADMMM算法(STEPD)来解决PSPI问题的宽松版本。关于比较基因监管网络的广泛案例研究(GNs)用来证明STEPD能够准确地推断结构突扰动和正常组织中的扰动,从而为计算生物学家提供了一种工具,用以识别新的预测性签名。一个光谱分析证实STEPDD可以恢复小型图层图变工具。
相关内容
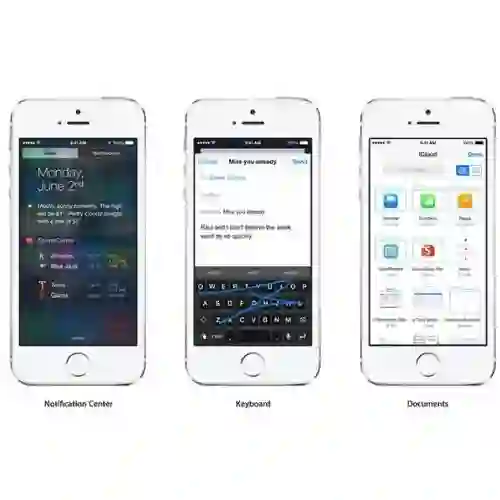
- Today (iOS and OS X): widgets for the Today view of Notification Center
- Share (iOS and OS X): post content to web services or share content with others
- Actions (iOS and OS X): app extensions to view or manipulate inside another app
- Photo Editing (iOS): edit a photo or video in Apple's Photos app with extensions from a third-party apps
- Finder Sync (OS X): remote file storage in the Finder with support for Finder content annotation
- Storage Provider (iOS): an interface between files inside an app and other apps on a user's device
- Custom Keyboard (iOS): system-wide alternative keyboards
Source: iOS 8 Extensions: Apple’s Plan for a Powerful App Ecosystem