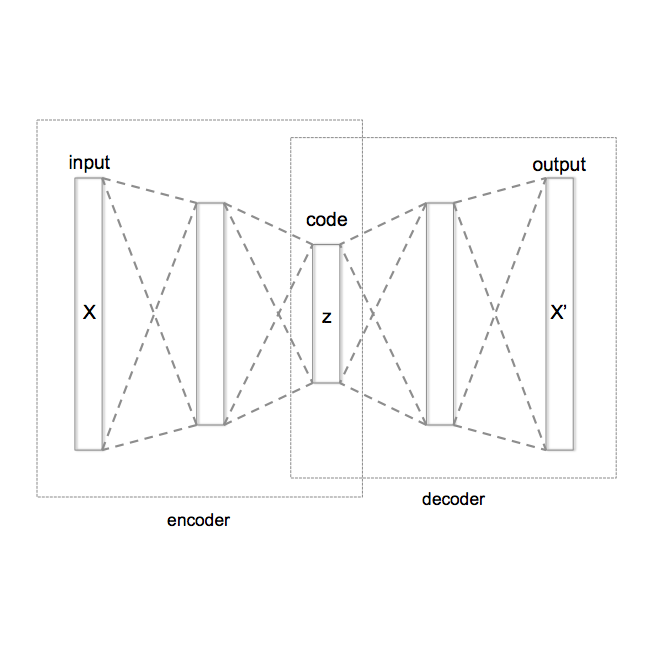
Bayesian optimisation (BO) is a standard approach for sample-efficient global optimisation of expensive black-box functions, yet its scalability to high dimensions remains challenging. Here, we investigate nonlinear dimensionality reduction techniques that reduce the problem to a sequence of low-dimensional Latent-Space BO (LSBO). While early LSBO methods used (linear) random projections (Wang et al., 2013), building on Grosnit et al. (2021), we employ Variational Autoencoders (VAEs) for LSBO, focusing on deep metric loss for structured latent manifolds and VAE retraining to adapt the encoder-decoder to newly sampled regions. We propose some changes in their implementation, originally designed for tasks such as molecule generation, and reformulate the algorithm for broader optimisation purposes. We then couple LSBO with Sequential Domain Reduction (SDR) directly in the latent space (SDR-LSBO), yielding an algorithm that narrows the latent search domains as evidence accumulates. Implemented in a GPU-accelerated BoTorch stack with Matern-5/2 Gaussian process surrogates, our numerical results show improved optimisation quality across benchmark tasks and that structured latent manifolds improve BO performance. Additionally, we compare random embeddings and VAEs as two mechanisms for dimensionality reduction, showing that the latter outperforms the former. To the best of our knowledge, this is the first study to combine SDR with VAE-based LSBO, and our analysis clarifies design choices for metric shaping and retraining that are critical for scalable latent space BO. For reproducibility, our source code is available at https://github.com/L-Lok/Nonlinear-Dimensionality-Reduction-Techniques-for-Bayesian-Optimization.git.
翻译:暂无翻译